diff --git a/docs/_images/anndata_schema.png b/docs/_images/anndata_schema.png
new file mode 100644
index 000000000..27f4b55aa
Binary files /dev/null and b/docs/_images/anndata_schema.png differ
diff --git a/docs/_rtd/data_types.md b/docs/_rtd/data_types.md
index b96dc73b0..15b83651e 100644
--- a/docs/_rtd/data_types.md
+++ b/docs/_rtd/data_types.md
@@ -40,8 +40,9 @@ For each cell, the following morphology features calculated from `skimage.measur
* `perimeter`: perimeter of object which approximates the contour as a line through the centers of border pixels using a 4-connectivity.
* `convex_area`: the area of the convex hull.
* `equivalent_diameter`: the diameter of the circle with the same area as the cell.
-* `centroid-0`: the $x$-coordinate of the centroid.
-* `centroid-1`: the $y$-coordinate of the centroid.
+* Centroids: Note that all the arrays are NumPy arrays, therefore the origin $(0,0)$ is in the "top-left corner" of the image / array.
+ * `centroid-0`: the $y$-coordinate of the centroid.
+ * `centroid-1`: the $x$-coordinate of the centroid.
* `fov`: The FOV from which the cell originates from.
The base `regionprops` metric often don't provide enough morphological information about each cell on their own. We add the following derived metrics to provide more complete information about the segmented cells:
@@ -49,7 +50,7 @@ The base `regionprops` metric often don't provide enough morphological informati
* `perim_square_over_area`: the square of the perimeter divided by the area.
* `major_axis_equiv_diam_ratio`: the major axis length divided by the equivalent diameter.
* `convex_hull_resid`: the difference between the convex area and the area divided by the convex area.
-* `centroid_dif`: the normalized euclidian distance between the cell centroid and the corresponding convex hull centroid.
+* `centroid_dif`: the normalized euclidean distance between the cell centroid and the corresponding convex hull centroid.
* `num_concavities`: the number of concavities of the region.
* `nc_ratio`: for nuclear segmentation only. The nuclear area divided by the total area.
@@ -90,3 +91,93 @@ The CSV should contain the following columns
* `fov`: name of the FOV the cell comes from
* `label`: the name of the segmentation label
* A set of expression columns defining the properties of each cell desired for clustering
+
+---
+
+Name: AnnData
+Type: anndata.AnnData
+Created by: [ConvertToAnnData](https://ark-analysis.readthedocs.io/en/latest/_markdown/ark.utils.html#ark.utils.data_utils.ConvertToAnnData)
+Used by: [anndata_conversion.ipynb](https://github.com/angelolab/ark-analysis/blob/main/templates/anndata_conversion.ipynb)
+
+
+  +
+
+
+
+`AnnData` is a data structure consisting of matrices, annotated by DataFrames and Indexes. The goal is to transition over to `AnnData` from the Cell Table as the primary tabular data structure for storing, and interacting with multiplexed spatial single cell data.
+This section will illustrate the components of the `AnnData` object, and provide brief examples of which cell table columns map to which `AnnData` components.
+
+A `AnnData` object is composed of the following components:
+
+- **X**
+- **var**
+- **obs**
+- **obsm**
+- **obsp**
+- **varm**
+- **varp**
+
+There will be one `AnnData` object per FOV. Each of these components have specific use cases and will be described below:
+
+### 1. X, var, obs
+
+
+  +
+
+
+- `X` is a matrix of shape `(n_obs, n_vars)` where `n_obs` is the number of observations (currently cell segmentations) and `n_vars` is the number of variables (currently number of channels / markers).
+
+For example the following columns from the Cell Table are mapped to the `X` component of the `AnnData` object:
+
+| CD14 | CD163 | CD20 | CD3 | CD31 | CD4 | CD45 | $\cdots$ | SMA | Vim |
+|----------|----------|----------|----------|----------|----------|----------|----------|----------|----------|
+| 0.1 | 0.3 | 0.4 | 0.1 | 0.3 | 0.1 | 0.1 | $\cdots$ | 0.4 | 0.8 |
+| $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ |
+| $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ |
+| 0.1 | 0.1 | 0.3 | 0.7 | 0.8 | 0.8 | 0.8 | $\cdots$ | 0.3 | 0.6 |
+
+- `var`: A `DataFrame` of shape `(..., n_vars)`, where the index is `var_names`. This `DataFrame` contains attributes of each variable. Currently, this goes unused as there is not a Cell Table analogue of this compartment, but in the future this may change.
+ - `var_names` is a `Pandas` Index where each value is a unique identifier for each variable. These are the names of the channels and should be unique.
+ - `n_vars` is the number of variables, and in this case it is the number of channels. Each channel is a variable, and each observation has a value for each channel.
+- `obs` A `DataFrame` of shape `(n_obs, ...)`, where the index is `obs_names`. This `DataFrame` contains information about each observation, such as numeric metrics from `regionprops` or categorical data such as cell phenotype, or patient-level information.
+ - `obs_names` is a `Pandas` Index where each value is a unique identifier for each observation. These are the names of the segmented regions, and should be unique.
+ - `n_obs` is the number of segmented regions or objects of interest. In this case, it is the number of segmented cells.
+
+For example, the following columns from the Cell Table are mapped to the `obs` component of the `AnnData` object:
+
+| label | area | eccentricity | $\cdots$ | centroid_dif | num_concavities | fov |
+|----------|----------|--------------|----------|--------------|-----------------|----------|
+| 1 | 345 | 0.2 | $\cdots$ | 0.01 | 0 | fov1 |
+| 2 | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ |
+| $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ | $\vdots$ |
+| 1112 | 460 | 0.11 | $\cdots$ | 0.1 | 12 | fov1 |
+
+
+
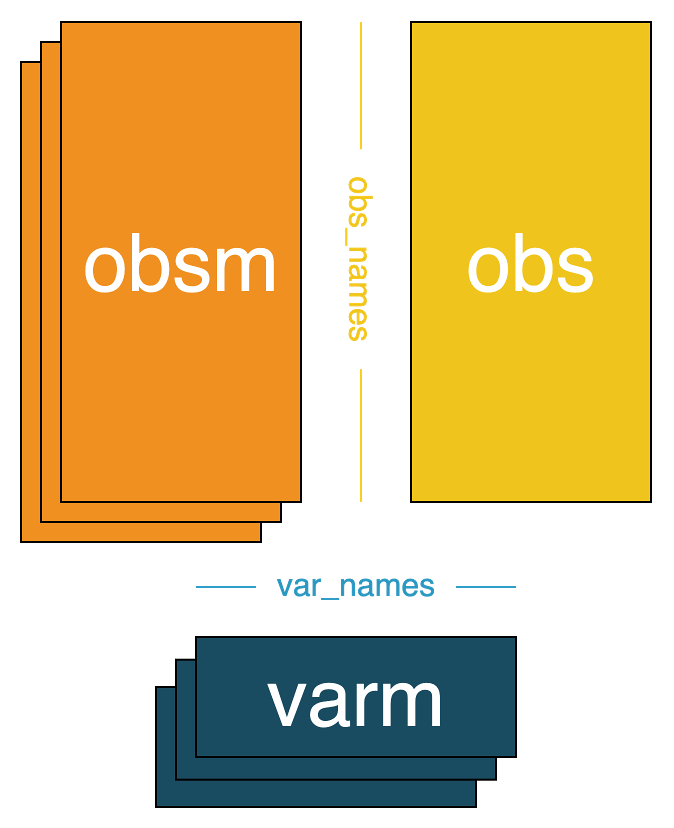
+### 2. obsm, varm
+
+
+  +
+
+
+- `obsm` is a key-value store where the values are matrices of shape `(n_obs, a)`, where `a` is an integer. This contains observation level matrices, and we use a mapping `str -> NDArray` to store them. For example, `"X_umap"` would store the UMAP embedding of the sparse matrix `X`, and `"X_pca"` would store the PCA embedding of `X`.
+ - Currently, from the Cell Table we store the `y` and `x` centroids in the `"spatial"` slot of the `obsm` component.
+- `varm` is a key-value store where the values are matrices of shape `(n_vars, b)`, where `b` is an integer. This contains variable level matrices, and we use a mapping `str -> NDArray` to store them. For example, `"Marker_umap"` would store the UMAP embedding of the matrix `var`.
+
+
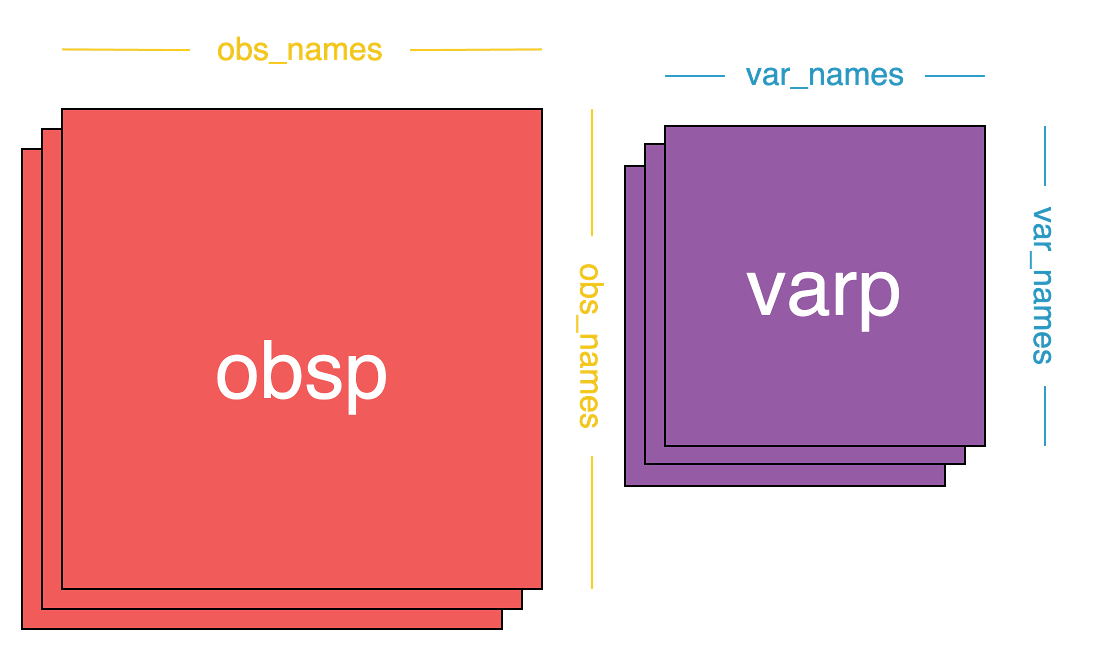
+### 3. obsp, varp
+
+
+  +
+
+
+- `obsp` is a square matrix of shape `(n_obs, n_obs)`, and its purpose is to store pairwise computations between observations.
+ - For example neighborhood information.
+- `varp` is a square matrix of shape `(n_vars, n_vars)`, and its purpose is to store pairwise computations between variables.
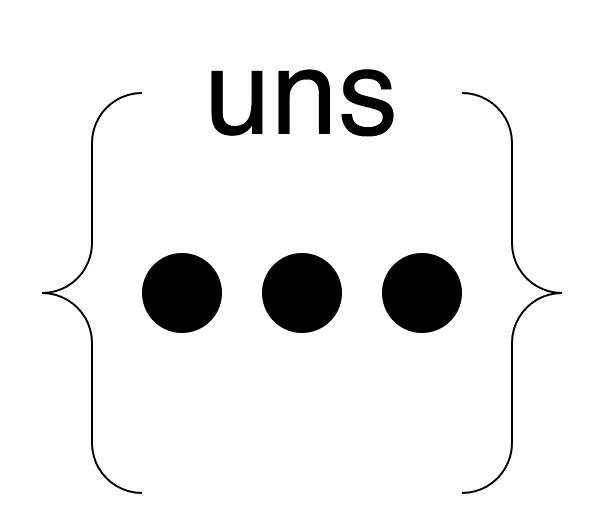
+### 4. **uns**
+
+
+  +
+
+
+- `uns` is a free slot for storing *almost* anything. It's a mapping from a string label to anything.
diff --git a/docs/_rtd/development.md b/docs/_rtd/development.md
index 1823f6f46..62276f3b5 100644
--- a/docs/_rtd/development.md
+++ b/docs/_rtd/development.md
@@ -144,3 +144,189 @@ Finally, to save an `xarray` to a file, use:
You can load the `xarray` back in using:
`arr = xr.load_dataarray(path)`
+
+
+### Working with `AnnData`
+
+We can load a single `AnnData` object using the function `anndata.read_zarr`, and several `AnnData` objects using the function `load_anndatas` from `ark.utils.data_utils`.
+
+```python
+from anndata import read_zarr
+from ark.utils.data_utils import load_anndatas
+```
+
+```python
+fov0 = read_zarr("data/example_dataset/fov0.zarr")
+```
+
+The channel intensities for each observation in the `AnnData` object with the `.to_df()` method, and get the channel names with `.var_names`.
+
+```python
+fov0.var_names
+fov0.to_df()
+```
+
+The observations and their properties with the `obs` property of the `AnnData` object. The data here consists of measurements such as `area`, `perimeter`, and categorical information like `cell_meta_cluster` for each cell.
+
+```python
+fov0.obs
+```
+
+The $x$ and $y$ centroids of each cell can be accessed with the `obsm` attribute and the key `"spatial"`.
+
+```python
+fov0.obsm["spatial"]
+```
+
+We can load all the `AnnData` objects in a directory lazily with `load_anndatas`. We get a view of the `AnnData` objects in the directory.
+
+```python
+fovs_ac = load_anndatas(anndata_dir = "data/example_dataset/fov0.zarr")
+```
+
+We can utilize `AnnData` objects or `AnnCollections` in a similar way to a Pandas DataFrame. For example, we can filter the `AnnCollection` to only include cells that have a `cell_meta_cluster` label of `"CD4T"`.
+
+```python
+fovs_ac_cd4t = fovs_ac[fovs_ac.obs["cell_meta_cluster"] == "CD4T"]
+print(type(fovs_ac_cd4t))
+fovs_ac_cd4t.obs.df
+```
+The type of `fovs_ac_cd4t` is not an `AnnData` object, but instead an `AnnCollectionView`.
+This is a `view` of the subset of the `AnnCollection`. This object can *only* access `.obs`, `.obsm`, `.layers` and `.X`.
+
+
+We can subset a `AnnCollectionView` to only include the first $n$ observations objects with the following code. The slice based indexing behaves like a `numpy` array.
+
+```python
+n = 100
+fovs_ac_cdt4_100 = fovs_ac_cd4t[:n]
+fovs_ac_cd4t_100.obs.df
+```
+
+Often we will want to subset the `AnnCollection` to only include observations contained within a specific FOV.
+
+```python
+fov1_adata = fovs_ac[fovs_ac.obs["fov"] == "fov1"]
+
+fov1_adata.obs.df
+```
+
+We can loop over all FOVs in a `AnnCollection` with the following code (there is alternative method in ):
+
+```python
+all_fovs = fovs_ac.obs["fov"].unique()
+
+for fov in all_fovs:
+ fov_adata = fovs_ac[fovs_ac.obs["fov"] == fov]
+ # do something with fov_adata
+```
+
+Functions which take in `AnnData` objects can often be applied to `AnnCollections`.
+
+The following works as expected:
+```python
+def dist(adata):
+ x = adata.obsm["spatial"]["centroid_x"]
+ y = adata.obsm["spatial"]["centroid_y"]
+ return np.sqrt(x**2 + y**2)
+
+dist(fovs_ac)
+```
+
+While the example below does not:
+```python
+from squidpy import gr
+gr.spatial_neighbors(adata=fovs_ac, spatial_key="spatial")
+```
+
+This is due to a `AnnCollection` object not having a `uns` property.
+
+#### Utilizing `DataLoaders`
+
+While a `AnnCollection` can sometimes be used to apply a function over all FOVs, in some instances we either cannot do that, or perhaps we want to apply functions to each FOV independently.
+
+We can access the underlying `AnnData` objects with `.adatas`.
+```python
+fovs_ac.adatas
+```
+
+In these instances we can construct data pipelines with [`torchdata`](https://pytorch.org/data/beta/index.html).
+
+
+As an example, let's create a multi-stage `DataLoader` which does the following:
+- Only extracts the observations with an area greater than `300`.
+- Only extracts the observations which have a `cell_meta_cluster` label of `"CD4T"`.
+- Compute the Spatial Neighbors graph for those observations (using [`squidpy.gr.spatial_neighbors`](https://squidpy.readthedocs.io/en/stable/api/squidpy.gr.spatial_neighbors.html#squidpy.gr.spatial_neighbors)).
+
+
+In order to construct a `torchdata` [`DataLoader2`](https://pytorch.org/data/beta/dataloader2.html) iterator we first need to create a `torchdata` [`IterDataPipe`](https://pytorch.org/data/beta/torchdata.datapipes.iter.html). This implements the `__iter__()` protocol, and represents an iterable over data samples.
+
+We can convert the `AnnCollection` to a `torchdata` `IterDataPipe` with `ark.utils.data_utils.AnnDataIterDataPipe`.
+
+
+```python
+from ark.utils.data_utils import AnnDataIterDataPipe
+
+fovs_ip = AnnDataIterDataPipe(fovs=fovs_ac)
+```
+
+The following two functions are used to filter the observations in the `AnnData` objects
+to only include cells with an area greater than `min_area` and cells with a `cell_meta_cluster` label of `cluster`.
+
+```python
+from anndata import AnnData
+
+def filter_cells_by_cluster(adata: AnnData, cluster_label: str) -> AnnData:
+ return adata[adata.obs["cell_meta_cluster"] == cluster_label]
+
+def filter_cells_by_area(adata: AnnData, min_area: int) -> AnnData:
+ return adata[adata.obs["area"] > min_area]
+```
+
+The following function is used to filter out `AnnData` objects which have no observations.
+```python
+def filter_empty_adata(fov: AnnData) -> bool:
+ return len(fov) > 0
+```
+
+
+We can apply these functions to the `IterDataPipe` with the `map` and the `filter` method.
+Because those methods return a new `IterDataPipe` object, we can chain them together.
+
+```python
+from functools import partial
+
+cd4t_obs_filter = partial(filter_cells_by_cluster, cluster_label="CD4T")
+area_obs_filter = partial(filter_cells_by_area, min_area=300)
+
+fovs_subset = fovs_ip.map(cd4t_obs_filter).map(area_obs_filter).filter(filter_empty_adata)
+```
+
+The data pipeline can be visualized with `to_graph` function.
+
+```python
+from torchdata.datapipes.utils import to_graph
+
+to_graph(fovs_subset)
+```
+
+
+The `DataLoader` can now be constructed.
+```python
+from torchdata.dataloader2.dataloader2 import DataLoader2
+
+fovs_subset_dl = DataLoader2(fovs_subset)
+```
+
+We can now loop over the `DataLoader` and compute the Spatial Neighbors graph per FOV with the filtered observations.
+
+```python
+for fov in fovs_subset_dl:
+ gr.spatial_neighbors(adata=fov, radius=350, spatial_key="spatial", coord_type="generic")
+```
+
+#### Further Reading
+- [Official AnnData Documentation](https://anndata.readthedocs.io/en/latest/)
+ - [Getting Started Tutorial](https://anndata.readthedocs.io/en/latest/tutorials/notebooks/getting-started.html)
+- [Converting from Single Cell Experiment and Seurat Objects](https://scanpy.readthedocs.io/en/stable/tutorials.html#conversion-anndata-singlecellexperiment-and-seurat-objects)
+- [MuData - Multimodal AnnData](https://mudata.readthedocs.io/en/latest/index.html)
diff --git a/docs/conf.py b/docs/conf.py
index 5154e7970..843b98e5a 100644
--- a/docs/conf.py
+++ b/docs/conf.py
@@ -72,6 +72,10 @@
'feather',
'google',
'h5py',
+ 'dask',
+ 'distributed',
+ 'anndata',
+ 'torchdata',
'ipywidgets',
'natsort',
'numba',
@@ -98,7 +102,8 @@
'mpl_toolkits',
'tqdm',
'ark.utils._bootstrapping',
- 'xmltodict']
+ 'xmltodict',
+ 'zarr',]
# prefix each section label with the name of the document it is in, followed by a colon
# autosection_label_prefix_document = True
diff --git a/pyproject.toml b/pyproject.toml
index 5a2ff3fd2..0fc7a5f8f 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -10,7 +10,9 @@ build-backend = "setuptools.build_meta"
[project]
dependencies = [
"alpineer==0.1.10",
+ "anndata",
"Cython>=0.29,<1",
+ "dask[distributed]",
"datasets>=2.6,<3.0",
"dill>=0.3.5,<0.4",
"feather-format>=0.4.1,<1",
@@ -29,16 +31,20 @@ dependencies = [
"requests>=2.20,<3",
"scikit-image<=0.19.3",
"scikit-learn>=1.1,<2",
+ "graphviz",
"scipy>=1.7,<2",
"seaborn>=0.12,<1",
"spatial-lda>=0.1.3,<1",
"statsmodels>=0.13.2,<1",
+ "squidpy",
"tifffile>=2022",
+ "torchdata",
"tqdm>=4,<5",
"umap-learn>=0.5,<1.0",
"xarray>=2022",
"xmltodict>=0.13.0,<1",
"zstandard>=0.19.0,<1",
+ "zarr",
"ark-analysis[colors]",
]
name = "ark-analysis"
diff --git a/src/ark/utils/data_utils.py b/src/ark/utils/data_utils.py
index e93a0736c..5afc6b32d 100644
--- a/src/ark/utils/data_utils.py
+++ b/src/ark/utils/data_utils.py
@@ -3,7 +3,8 @@
import os
import pathlib
import re
-from typing import List, Union
+from typing import List, Literal, Union, Sequence
+
from numpy.typing import ArrayLike, DTypeLike
from numpy import ma
import feather
@@ -17,6 +18,19 @@
import xarray as xr
from ark import settings
from skimage.segmentation import find_boundaries
+import dask.dataframe as dd
+from dask import delayed
+from anndata import AnnData, read_zarr
+from anndata.experimental import AnnCollection
+from anndata.experimental.multi_files._anncollection import ConvertType
+from tqdm.dask import TqdmCallback
+from torchdata.datapipes.iter import IterDataPipe
+from typing import Iterator, Optional
+try:
+ from typing import TypedDict, Unpack
+except ImportError:
+ from typing_extensions import TypedDict, Unpack
+
def save_fov_mask(fov, data_dir, mask_data, sub_dir=None, name_suffix=''):
@@ -177,7 +191,6 @@ def fov_mapping(self, fov: str) -> pd.DataFrame:
"""
misc_utils.verify_in_list(requested_fov=[fov], all_fovs=self.unique_fovs)
fov_data: pd.DataFrame = self.mapping[self.mapping[self.fov_column] == fov]
-
return fov_data.reset_index(drop=True)
@property
@@ -805,3 +818,219 @@ def stitch_images_by_shape(data_dir, stitched_dir, img_sub_folder=None, channels
current_img = stitched_data.loc['stitched_image', :, :, chan].values
image_utils.save_image(os.path.join(stitched_subdir, chan + '_stitched' + file_ext),
current_img)
+
+
+@delayed
+def _convert_ct_fov_to_adata(fov_dd: dd.DataFrame, var_names: list[str], obs_names: list[str], save_dir: os.PathLike) -> str:
+ """Converts the cell table for a single FOV to an `AnnData` object and saves it to disk as a
+ `Zarr` store.
+
+ Parameters
+ ----------
+ fov_dd : dd.DataFrame
+ The cell table subset on a single FOV.
+ var_names: list[str]
+ The marker names to extract from the cell table.
+ obs_names: list[str]
+ The cell-level measurements and properties to extract from the cell table.
+ save_dir: os.PathLike
+ The directory to save the `AnnData` object to.
+
+ Returns
+ -------
+ str
+ The path of the saved `AnnData` object.
+ """
+
+ fov_dd: dd.DataFrame = fov_dd.sort_values(by=settings.CELL_LABEL, key=ns.natsort_key).reset_index()
+ fov_id: str = fov_dd[settings.FOV_ID].iloc[0]
+
+ # Set the index to be the FOV and the segmentation label to create a unique index
+ fov_dd.index = list(map(lambda label: f"{fov_id}_{int(label)}", fov_dd[settings.CELL_LABEL]))
+
+ # Extract the X matrix
+ X_dd: dd.DataFrame = fov_dd[var_names]
+
+ # Extract the obs dataframe and convert the cell label to integer
+ obs_dd: dd.DataFrame = fov_dd[obs_names].astype({settings.CELL_LABEL: int, settings.FOV_ID: str})
+ obs_dd["cell_meta_cluster"] = pd.Categorical(obs_dd["cell_meta_cluster"].astype(str))
+
+ # Move centroids from obs to obsm["spatial"]
+ obsm_dd = obs_dd[[settings.CENTROID_0, settings.CENTROID_1]].rename(columns={settings.CENTROID_0: "centroid_y", settings.CENTROID_1: "centroid_x"})
+ obs_dd = obs_dd.drop(columns=[settings.CENTROID_0, settings.CENTROID_1])
+
+ # Create the AnnData object
+ adata: AnnData = AnnData(X=X_dd, obs=obs_dd, obsm={"spatial": obsm_dd})
+
+ # Convert any extra string labels to categorical if it's beneficial.
+ adata.strings_to_categoricals()
+
+ adata.write_zarr(pathlib.Path(save_dir, f"{fov_id}.zarr"), chunks=(1000, 1000))
+ return pathlib.Path(save_dir, f"{fov_id}.zarr").as_posix()
+
+
+class ConvertToAnnData:
+ """ A class which converts the Cell Table `.csv` file to a series of `AnnData` objects,
+ one object per FOV.
+
+ The default parameters stored in the `.obs` slot include:
+ - `area`
+ - `cell_meta_cluster`
+ - `centroid_dif`
+ - `convex_area`
+ - `convex_hull_resid`
+ - `cell_meta_cluster`
+ - `eccentricity`
+ - `fov`
+ - `major_axis_equiv_diam_ratio`
+
+ Visit the Data Types document to see the full list of parameters.
+ The default parameters stored in the `.obs` slot include:
+ - `centroid_x`
+ - `centroid_y`
+
+ Args:
+ cell_table_path (os.PathLike): The path to the cell table.
+ markers (list[str], "auto"): The markers to extract and store in `.X`. Defaults to "auto",
+ which will extract all markers.
+ extra_obs_parameters (list[str], optional): Extra parameters to load in `.obs`. Defaults
+ to None.
+ """
+
+ def __init__(self, cell_table_path: os.PathLike,
+ markers: Union[list[str], Literal["auto"]] = "auto",
+ extra_obs_parameters: list[str] = None) -> None:
+
+ io_utils.validate_paths(paths=cell_table_path)
+
+
+ # Read in the cell table
+ cell_table: dd.DataFrame = dd.read_csv(cell_table_path)
+ ct_columns = cell_table.columns
+
+ # Get the marker column indices
+ marker_index_start: int = ct_columns.get_loc(settings.PRE_CHANNEL_COL) + 1
+ marker_index_stop: int = ct_columns.get_loc(settings.POST_CHANNEL_COL)
+ obs_index_start: int = ct_columns.get_loc(settings.POST_CHANNEL_COL) + 1
+
+ if markers == "auto":
+ # Default to all markers based on settings Pre and Post channel column values
+ markers: list[str] = ct_columns[marker_index_start:marker_index_stop].to_list()
+ else:
+ # Verify that the correct markers exist
+ misc_utils.verify_in_list(requested_markers=markers,
+ all_markers=ct_columns[marker_index_start:marker_index_stop].to_list())
+ self.var_names = markers
+
+ # Verify extra obs parameters
+ if extra_obs_parameters:
+ misc_utils.verify_in_list(requested_parameters=extra_obs_parameters,
+ all_parameters=ct_columns[obs_index_start:].to_list())
+ else:
+ extra_obs_parameters = []
+
+ obs_names = [
+ settings.CELL_LABEL,
+ settings.CELL_SIZE,
+ *ct_columns[obs_index_start:].to_list(),
+ *extra_obs_parameters
+ ]
+
+ # Use "area" as the default area id instead of settings.CELL_SIZE to account for
+ # non-cellular observations (ez_seg, fiber, etc...)
+ if settings.CELL_SIZE in obs_names:
+ obs_names.remove(settings.CELL_SIZE)
+ if "area" not in obs_names:
+ cell_table = cell_table.rename(columns={settings.CELL_SIZE: "area"})
+ obs_names.append("area")
+
+ self.obs_names: list[str] = obs_names
+ self.cell_table = cell_table
+
+ def convert_to_adata(
+ self,
+ save_dir: os.PathLike,
+ ) -> dict[str, str]:
+ """Converts the cell table to a FOV-level `AnnData` object, and saves the results as
+ a `Zarr` store to disk in the `save_dir`.
+
+ Args:
+ save_dir (os.PathLike): The directory to save the `AnnData` objects to.
+
+ Returns:
+ dict[str, str]: A dictionary containing the names of the FOVs and the paths where
+ they were saved.
+ """
+
+ if not isinstance(save_dir, pathlib.Path):
+ save_dir = pathlib.Path(save_dir)
+ if not save_dir.exists():
+ save_dir.mkdir(parents=True, exist_ok=True)
+
+
+ with TqdmCallback(desc="Converting to AnnData"):
+ g: pd.Series = (
+ self.cell_table.groupby(by=settings.FOV_ID, sort=True)
+ .apply(
+ _convert_ct_fov_to_adata,
+ var_names=self.var_names,
+ obs_names=self.obs_names,
+ save_dir=save_dir,
+ meta=("anndata_save_results", str),
+ )
+ ).compute()
+
+ return g.to_dict()
+
+
+class AnnCollectionKwargs(TypedDict):
+ join_obs: Optional[Literal["inner", "outer"]]
+ join_obsm: Optional[Literal["inner"]]
+ join_vars: Optional[Literal["inner"]]
+ label: Optional[str]
+ keys: Optional[Sequence[str]]
+ index_unique: Optional[str]
+ convert: Optional[ConvertType]
+ harmonize_dtypes: bool
+ indices_strict: bool
+
+
+def load_anndatas(anndata_dir: os.PathLike, **anncollection_kwargs: Unpack[AnnCollectionKwargs]) -> AnnCollection:
+ """Lazily loads a directory of `AnnData` objects into an `AnnCollection`. The concatination happens across the `.obs` axis.
+
+ For `AnnCollection` kwargs, see https://anndata.readthedocs.io/en/latest/generated/anndata.experimental.AnnCollection.html
+
+ Args:
+ anndata_dir (os.PathLike): The directory containing the `AnnData` objects.
+
+ Returns:
+ AnnCollection: The `AnnCollection` containing the `AnnData` objects.
+ """
+ if not isinstance(anndata_dir, pathlib.Path):
+ anndata_dir = pathlib.Path(anndata_dir)
+
+ adata_zarr_stores = {f.stem: read_zarr(f) for f in ns.natsorted(anndata_dir.glob("*.zarr"))}
+ return AnnCollection(adatas=adata_zarr_stores, **anncollection_kwargs)
+
+
+class AnnDataIterDataPipe(IterDataPipe):
+ """The TorchData Iterable-style DataPipe. Takes an `AnnCollection`
+ and makes it iterable by FOV for easy and flexible data pipelines.
+
+ Args:
+ fovs (AnnCollection): The `AnnCollection` containing the `AnnData` objects.
+ """
+
+ @property
+ def fovs(self) -> AnnCollection:
+ return self._fovs
+
+ @fovs.setter
+ def fovs(self, value: AnnCollection) -> None:
+ self._fovs: AnnCollection = value
+
+ def __init__(self, fovs: AnnCollection):
+ self.fovs = fovs
+
+ def __iter__(self) -> Iterator[AnnData]:
+ yield from self.fovs.adatas
diff --git a/templates/anndata_conversion.ipynb b/templates/anndata_conversion.ipynb
new file mode 100644
index 000000000..bbee809e1
--- /dev/null
+++ b/templates/anndata_conversion.ipynb
@@ -0,0 +1,207 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# `AnnData` Conversion\n",
+ "\n",
+ "The purpose of this notebook is to convert the cell table to a [`AnnData`](https://anndata.readthedocs.io/en/latest/index.html) Object.\n",
+ "\n",
+ "`AnnData` stands for Annotated Data, and is a data structure well suited for single cell data. It is a multi-faceted object composed of matrices and DataFrames which can be used to efficiently store and interact with our data.\n",
+ "\n",
+ "The following is a representation of the `AnnData` object schema:\n",
+ "\n",
+ "\n",
+ "  \n",
+ "
\n",
+ "
\n",
+ "\n",
+ "This notebook will move the following portions of the Cell Table to a `AnnData` object:\n",
+ "- Markers / Channel columns get stored in `.X`\n",
+ "- The X and Y Centroids get stored in `.obs`\n",
+ "- The rest of the cell table gets stored in `.obs` (includes columns such as `area`, `perimeter`, `cell_meta_cluster`)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "ExecuteTime": {
+ "end_time": "2023-11-30T17:09:00.527535Z",
+ "start_time": "2023-11-30T17:08:56.476970Z"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "from dask.distributed import Client\n",
+ "from anndata import read_zarr\n",
+ "from ark.utils.data_utils import ConvertToAnnData\n",
+ "import os"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "ExecuteTime": {
+ "end_time": "2023-11-30T17:09:02.298445Z",
+ "start_time": "2023-11-30T17:09:00.528933Z"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "Client(threads_per_worker = 2)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "ExecuteTime": {
+ "end_time": "2023-11-30T17:08:54.069774Z",
+ "start_time": "2023-11-30T17:08:54.067066Z"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "base_dir = \"../data/example_dataset/\""
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "## 0. Download the Example Dataset\n",
+ "\n",
+ "Here we are using the example data located in `/data/example_dataset/input_data/`. To modify this notebook to run using your own data, simply change `base_dir` to point to your own sub-directory within the data folder.\n",
+ "\n",
+ "* `base_dir`: the path to all of your imaging data. This directory will contain all of the data generated by this notebook, as well as the data previously generated by segmentation and cell clustering."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "ExecuteTime": {
+ "end_time": "2023-11-30T17:09:04.139994Z",
+ "start_time": "2023-11-30T17:09:02.283610Z"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "from ark.utils.example_dataset import get_example_dataset\n",
+ "\n",
+ "get_example_dataset(dataset=\"post_clustering\", save_dir= base_dir, overwrite_existing=True)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {
+ "collapsed": false
+ },
+ "source": [
+ "## 1. Convert the Cell Table to `AnnData` Objects\n",
+ "\n",
+ "- `cell_table_path`: The path to the cell table that you wish to convert to `AnnData` objects. \n",
+ "- `anndata_save_dir`: The directory where you would like to save the `AnnData` objects. This directory will be created if it does not already exist."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "ExecuteTime": {
+ "end_time": "2023-11-30T17:09:55.614306Z",
+ "start_time": "2023-11-30T17:09:55.607681Z"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "cell_table_path = os.path.join(base_dir, \"segmentation/cell_table/cell_table_size_normalized_cell_labels.csv\")\n",
+ "anndata_save_dir = os.path.join(base_dir, \"anndata\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "- `markers`: These are the names of the markers that you wish to extract from the Cell Table. You can specify each marker that you would like to use, or you may set it to `\"auto\"` in order to grab all markers.\n",
+ "- `extra_obs_parameters`: By default the conversion extracts a specific set of columns for the `obs` DataFrame, and all columns to the left of `\"label\"`. If you would like to add additional columns to the `obs` DataFrame, you can specify them with this parameter."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "ExecuteTime": {
+ "end_time": "2023-11-30T17:09:56.466954Z",
+ "start_time": "2023-11-30T17:09:56.464196Z"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "# markers = [\"CD14\", \"CD163\", \"CD20\", \"CD3\", \"CD31\", \"CD4\", \"CD45\", \"CD68\", \"CD8\", \"CK17\", \"Collagen1\", \"ECAD\",\n",
+ "# \"Fibronectin\", \"GLUT1\", \"H3K27me3\", \"H3K9ac\", \"HLADR\", \"IDO\", \"Ki67\", \"PD1\", \"SMA\", \"Vim\"]\n",
+ "markers = \"auto\"\n",
+ "extra_obs_parameters = None"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "ExecuteTime": {
+ "end_time": "2023-11-30T17:09:56.879793Z",
+ "start_time": "2023-11-30T17:09:56.867649Z"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "convert_to_anndata = ConvertToAnnData(cell_table_path, markers=markers, extra_obs_parameters=extra_obs_parameters)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "ExecuteTime": {
+ "end_time": "2023-11-30T17:10:01.796185Z",
+ "start_time": "2023-11-30T17:09:57.186645Z"
+ },
+ "collapsed": false
+ },
+ "outputs": [],
+ "source": [
+ "fov_adata_paths = convert_to_anndata.convert_to_adata(save_dir=anndata_save_dir)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "We recommend reading both a brief overview of the `AnnData` datatype documentation [here](https://ark-analysis.readthedocs.io/en/latest/_rtd/data_types.html), and the official documentation [here](https://anndata.readthedocs.io/en/latest/index.html)."
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "ark",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.11.5"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 2
+}
diff --git a/tests/analysis/dimensionality_reduction_test.py b/tests/analysis/dimensionality_reduction_test.py
index 30c784e7c..472769250 100644
--- a/tests/analysis/dimensionality_reduction_test.py
+++ b/tests/analysis/dimensionality_reduction_test.py
@@ -10,7 +10,7 @@
def test_plot_dim_reduced_data():
# this only tests errors, test_dimensionality_reduction tests the meat of this function
- random_cell_data = test_utils.make_cell_table(50)
+ random_cell_data = test_utils.make_cell_table(n_cells=50, n_markers=10)
with pytest.raises(FileNotFoundError):
# trying to save to a non-existant directory
@@ -34,8 +34,9 @@ def test_plot_dim_reduced_data():
def test_dimensionality_reduction():
- random_cell_data = test_utils.make_cell_table(50)
- test_cols = test_utils.TEST_MARKERS
+ n_markers = 4
+ random_cell_data = test_utils.make_cell_table(n_cells=50, n_markers=n_markers)
+ test_cols = [f"marker_{i}" for i in range(n_markers)]
test_algorithms = ['PCA', 'tSNE', 'UMAP']
diff --git a/tests/analysis/visualize_test.py b/tests/analysis/visualize_test.py
index 9f19d47a2..bdd502c24 100644
--- a/tests/analysis/visualize_test.py
+++ b/tests/analysis/visualize_test.py
@@ -6,10 +6,12 @@
import pytest
import test_utils
import xarray as xr
+import pandas as pd
import ark.settings as settings
import ark.spLDA.processing as pros
from ark.analysis import visualize
+import test_utils as ark_test_utils
def test_draw_heatmap():
@@ -55,72 +57,78 @@ def test_draw_heatmap():
assert os.path.exists(os.path.join(temp_dir, "z_score_viz.png"))
-def test_draw_boxplot():
+@pytest.fixture
+def test_cell_table() -> pd.DataFrame:
+ n_cells = 300
+ n_markers = 5
+ cell_table = ark_test_utils.make_cell_table(n_cells=n_cells, n_markers=n_markers)
+ yield cell_table
+
+
+def test_draw_boxplot(test_cell_table: pd.DataFrame):
# trim random data so we don't have to visualize as many facets
- start_time = timeit.default_timer()
- random_data = test_utils.make_cell_table(100)
- random_data = random_data[random_data[settings.PATIENT_ID].isin(np.arange(1, 5))]
+ random_data = test_cell_table[test_cell_table[settings.PATIENT_ID].isin(np.arange(1, 5))]
# basic error testing
with pytest.raises(ValueError):
# non-existant col_name
- visualize.draw_boxplot(cell_data=random_data, col_name="AA")
+ visualize.draw_boxplot(cell_data=random_data, col_name="bad_marker")
with pytest.raises(ValueError):
# split_vals specified but not col_split
- visualize.draw_boxplot(cell_data=random_data, col_name="A", split_vals=[])
+ visualize.draw_boxplot(cell_data=random_data, col_name="marker_1", split_vals=[])
with pytest.raises(ValueError):
# non-existant col_split specified
- visualize.draw_boxplot(cell_data=random_data, col_name="A", col_split="AA")
+ visualize.draw_boxplot(cell_data=random_data, col_name="marker_1", col_split="AA")
with pytest.raises(ValueError):
# split_vals not found in col_split found
- visualize.draw_boxplot(cell_data=random_data, col_name="A",
+ visualize.draw_boxplot(cell_data=random_data, col_name="marker_1",
col_split=settings.PATIENT_ID, split_vals=[3, 4, 5, 6])
with pytest.raises(FileNotFoundError):
# trying to save to a non-existant directory
- visualize.draw_boxplot(cell_data=random_data, col_name="A",
+ visualize.draw_boxplot(cell_data=random_data, col_name="marker_1",
save_dir="bad_dir")
# highest level: data, a column name, a split column, and split vals
with tempfile.TemporaryDirectory() as temp_dir:
- visualize.draw_boxplot(cell_data=random_data, col_name="A",
+ visualize.draw_boxplot(cell_data=random_data, col_name="marker_1",
col_split=settings.PATIENT_ID, split_vals=[1, 2],
save_dir=temp_dir, save_file="boxplot_viz.png")
assert os.path.exists(os.path.join(temp_dir, "boxplot_viz.png"))
-def test_get_sort_data():
- random_data = test_utils.make_cell_table(100)
- sorted_data = visualize.get_sorted_data(random_data, settings.PATIENT_ID, settings.CELL_TYPE)
+def test_get_sort_data(test_cell_table: pd.DataFrame):
+ sorted_data = visualize.get_sorted_data(
+ test_cell_table,
+ settings.PATIENT_ID,
+ settings.CELL_TYPE
+ )
row_sums = [row.sum() for index, row in sorted_data.iterrows()]
assert list(reversed(row_sums)) == sorted(row_sums)
-def test_plot_barchart():
+def test_plot_barchart(test_cell_table):
# mostly error checking here, test_visualize_cells tests the meat of the functionality
- random_data = test_utils.make_cell_table(100)
with pytest.raises(FileNotFoundError):
# trying to save to a non-existant directory
- visualize.plot_barchart(random_data, "Random Title", "Random X Label",
+ visualize.plot_barchart(test_cell_table, "Random Title", "Random X Label",
"Random Y Label", save_dir="bad_dir")
with pytest.raises(FileNotFoundError):
# setting save_dir but not setting save_file
- visualize.plot_barchart(random_data, "Random Title", "Random X Label",
+ visualize.plot_barchart(test_cell_table, "Random Title", "Random X Label",
"Random Y Label", save_dir=".")
-def test_visualize_patient_population_distribution():
- random_data = test_utils.make_cell_table(100)
-
+def test_visualize_patient_population_distribution(test_cell_table):
with tempfile.TemporaryDirectory() as temp_dir:
# test without a save_dir, check that we do not save the files
- visualize.visualize_patient_population_distribution(random_data, settings.PATIENT_ID,
+ visualize.visualize_patient_population_distribution(test_cell_table, settings.PATIENT_ID,
settings.CELL_TYPE)
assert not os.path.exists(os.path.join(temp_dir, "PopulationDistribution.png"))
@@ -128,7 +136,7 @@ def test_visualize_patient_population_distribution():
assert not os.path.exists(os.path.join(temp_dir, "PopulationProportion.png"))
# now test with a save_dir, which will check that we do save the files
- visualize.visualize_patient_population_distribution(random_data, settings.PATIENT_ID,
+ visualize.visualize_patient_population_distribution(test_cell_table, settings.PATIENT_ID,
settings.CELL_TYPE, save_dir=temp_dir)
# Check if correct plots are saved
@@ -162,11 +170,10 @@ def test_visualize_neighbor_cluster_metrics():
assert os.path.exists(os.path.join(temp_dir, "neighborhood_silhouette_scores.png"))
-def test_visualize_topic_eda():
+def test_visualize_topic_eda(test_cell_table: pd.DataFrame):
# Create/format/featurize testing cell table
- cell_table = test_utils.make_cell_table(num_cells=1000)
- all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
- cell_table_format = pros.format_cell_table(cell_table, clusters=all_clusters)
+ all_clusters = list(np.unique(test_cell_table[settings.CELL_TYPE]))
+ cell_table_format = pros.format_cell_table(test_cell_table, clusters=all_clusters)
cell_table_features = pros.featurize_cell_table(cell_table_format)
# Run topic EDA
@@ -201,11 +208,10 @@ def test_visualize_topic_eda():
"topic_eda_cell_counts_k_{}.png".format(tops[0])))
-def test_visualize_fov_stats():
+def test_visualize_fov_stats(test_cell_table: pd.DataFrame):
# Create/format/featurize testing cell table
- cell_table = test_utils.make_cell_table(num_cells=1000)
- all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
- cell_table_format = pros.format_cell_table(cell_table, clusters=all_clusters)
+ all_clusters = list(np.unique(test_cell_table[settings.CELL_TYPE]))
+ cell_table_format = pros.format_cell_table(test_cell_table, clusters=all_clusters)
# Run topic EDA
fov_stats = pros.fov_density(cell_table_format)
@@ -227,10 +233,9 @@ def test_visualize_fov_stats():
assert os.path.exists(os.path.join(temp_dir, "fov_metrics_total_cells.png"))
-def test_visualize_fov_graphs():
- cell_table = test_utils.make_cell_table(num_cells=1000)
- all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
- cell_table_format = pros.format_cell_table(cell_table, clusters=all_clusters)
+def test_visualize_fov_graphs(test_cell_table: pd.DataFrame):
+ all_clusters = list(np.unique(test_cell_table[settings.CELL_TYPE]))
+ cell_table_format = pros.format_cell_table(test_cell_table, clusters=all_clusters)
cell_table_features = pros.featurize_cell_table(cell_table_format)
diff_mats = pros.create_difference_matrices(cell_table_format, cell_table_features)
diff --git a/tests/segmentation/segmentation_utils_test.py b/tests/segmentation/segmentation_utils_test.py
index 8aa2d63f8..4b69ff374 100644
--- a/tests/segmentation/segmentation_utils_test.py
+++ b/tests/segmentation/segmentation_utils_test.py
@@ -290,8 +290,8 @@ def test_save_segmentation_labels():
def test_concatenate_csv():
# create sample data
- test_data_1 = test_utils.make_cell_table(num_cells=10)
- test_data_2 = test_utils.make_cell_table(num_cells=20)
+ test_data_1 = test_utils.make_cell_table(n_cells=10, n_markers=5)
+ test_data_2 = test_utils.make_cell_table(n_cells=20, n_markers=5)
with pytest.raises(ValueError):
# attempt to pass column_values list with different length than number of csv files
diff --git a/tests/spLDA/processing_test.py b/tests/spLDA/processing_test.py
index a91af404d..42ad2ef74 100644
--- a/tests/spLDA/processing_test.py
+++ b/tests/spLDA/processing_test.py
@@ -3,28 +3,38 @@
from alpineer import misc_utils
from sklearn.cluster import KMeans
from test_utils import make_cell_table
-
+from typing import List, Tuple, Callable
+import pandas as pd
import ark.settings as settings
import ark.spLDA.processing as pros
from ark.utils.spatial_lda_utils import within_cluster_sums
-# Generate a test cell table
-N_CELLS = 1000
-TEST_CELL_TABLE = make_cell_table(N_CELLS)
+
+@pytest.fixture(scope="module")
+def test_cell_table() -> Callable:
+ def generate_cell_table(n_cells: int, n_markers: int) -> Tuple[pd.DataFrame, List[str]]:
+ n_markers = 10
+ n_cells = 1000
+ cell_table = make_cell_table(n_cells=n_cells, n_markers=n_markers)
+ marker_names = [f"marker_{i}" for i in range(n_markers)]
+ return cell_table, marker_names
+ yield generate_cell_table
-def test_format_cell_table():
+def test_format_cell_table(test_cell_table: Callable):
# call formatting function
- all_clusters = list(np.unique(TEST_CELL_TABLE[settings.CELL_TYPE]))
- all_markers = ['A', 'B', 'C', 'D', 'E', 'F', 'G']
+ n_markers = 10
+ n_cells = 1000
+ cell_table, all_markers = test_cell_table(n_cells=n_cells, n_markers=n_markers)
+ all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
some_clusters = all_clusters[2:]
some_markers = all_markers[2:]
- all_clusters_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE, clusters=all_clusters)
- all_markers_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE, markers=all_markers)
- some_clusters_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE,
+ all_clusters_format = pros.format_cell_table(cell_table=cell_table, clusters=all_clusters)
+ all_markers_format = pros.format_cell_table(cell_table=cell_table, markers=all_markers)
+ some_clusters_format = pros.format_cell_table(cell_table=cell_table,
clusters=some_clusters)
- some_markers_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE, markers=some_markers)
+ some_markers_format = pros.format_cell_table(cell_table=cell_table, markers=some_markers)
# Check that number of FOVS match
cluster_fovs = [x for x in all_clusters_format.keys() if
@@ -32,9 +42,9 @@ def test_format_cell_table():
marker_fovs = [x for x in all_markers_format.keys() if
x not in ['fovs', 'markers', 'clusters']]
misc_utils.verify_in_list(
- fovs1=list(np.unique(TEST_CELL_TABLE[settings.FOV_ID])), fovs2=cluster_fovs)
+ fovs1=list(np.unique(cell_table[settings.FOV_ID])), fovs2=cluster_fovs)
misc_utils.verify_in_list(
- fovs1=list(np.unique(TEST_CELL_TABLE[settings.FOV_ID])), fovs2=marker_fovs)
+ fovs1=list(np.unique(cell_table[settings.FOV_ID])), fovs2=marker_fovs)
# Check that columns were retained/renamed
misc_utils.verify_in_list(
@@ -45,20 +55,21 @@ def test_format_cell_table():
cols2=list(all_markers_format[1].columns))
# Check that columns were dropped
- assert len(TEST_CELL_TABLE.columns) > len(all_clusters_format[1].columns)
- assert len(TEST_CELL_TABLE.columns) > len(all_markers_format[1].columns)
+ assert len(cell_table.columns) > len(all_clusters_format[1].columns)
+ assert len(cell_table.columns) > len(all_markers_format[1].columns)
# check that only specified clusters and markers are kept
assert not np.isin(all_clusters[:2], np.unique(some_clusters_format[1].cluster)).any()
assert not np.isin(all_markers[:2], np.unique(some_markers_format[1].columns)).any()
-def test_featurize_cell_table():
- # call formatting function
- all_clusters = list(np.unique(TEST_CELL_TABLE[settings.CELL_TYPE]))
- all_markers = ['A', 'B', 'C', 'D', 'E', 'F', 'G']
- cluster_names = list(np.unique(TEST_CELL_TABLE[settings.CELL_TYPE]))
- cell_table_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE, clusters=all_clusters,
+def test_featurize_cell_table(test_cell_table: Callable):
+ n_markers = 10
+ n_cells = 1000
+ cell_table, all_markers = test_cell_table(n_cells=n_cells, n_markers=n_markers)
+
+ all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
+ cell_table_format = pros.format_cell_table(cell_table=cell_table, clusters=all_clusters,
markers=all_markers)
# call featurization on different training fractions
@@ -73,23 +84,26 @@ def test_featurize_cell_table():
train_frac=0.75)
# Check for consistent dimensions and correct column names
- assert all_clusters_75["featurized_fovs"].shape[0] == TEST_CELL_TABLE.shape[0] == N_CELLS
- assert all_clusters_50["featurized_fovs"].shape[0] == TEST_CELL_TABLE.shape[0] == N_CELLS
- assert all_clusters_75["train_features"].shape[0] == 0.75 * N_CELLS
- assert all_clusters_50["train_features"].shape[0] == 0.5 * N_CELLS
+ assert all_clusters_75["featurized_fovs"].shape[0] == cell_table.shape[0] == n_cells
+ assert all_clusters_50["featurized_fovs"].shape[0] == cell_table.shape[0] == n_cells
+ assert all_clusters_75["train_features"].shape[0] == 0.75 * n_cells
+ assert all_clusters_50["train_features"].shape[0] == 0.5 * n_cells
misc_utils.verify_in_list(correct=all_markers,
actual=list(all_markers_75["featurized_fovs"].columns))
- misc_utils.verify_in_list(correct=cluster_names,
+ misc_utils.verify_in_list(correct=all_clusters,
actual=list(all_clusters_75["featurized_fovs"].columns))
# check for correct featurization method
assert all_clusters_75["featurization"] == "cluster"
assert all_markers_75["featurization"] == "marker"
-def test_gap_stat():
- # call formatting & featurization - only test on clusters to avoid repetition
- all_clusters = list(np.unique(TEST_CELL_TABLE[settings.CELL_TYPE]))
- all_clusters_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE, clusters=all_clusters)
+def test_gap_stat(test_cell_table: Callable):
+ n_markers = 10
+ n_cells = 1000
+ cell_table, all_markers = test_cell_table(n_cells=n_cells, n_markers=n_markers)
+ all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
+
+ all_clusters_format = pros.format_cell_table(cell_table=cell_table, clusters=all_clusters)
features = pros.featurize_cell_table(cell_table=all_clusters_format, featurization='cluster')
clust_labs = KMeans(n_clusters=5).fit(features['featurized_fovs']).labels_
clust_sums = within_cluster_sums(features['featurized_fovs'], clust_labs)
@@ -104,10 +118,13 @@ def test_gap_stat():
assert gap[0] >= 0 and gap[1] >= 0
-def test_compute_topic_eda():
- # Format & featurize cell table. Only test on clusters and 0.75 train frac to avoid repetition
- all_clusters = list(np.unique(TEST_CELL_TABLE[settings.CELL_TYPE]))
- all_clusters_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE, clusters=all_clusters)
+def test_compute_topic_eda(test_cell_table: Callable):
+ n_markers = 10
+ n_cells = 1000
+ cell_table, all_markers = test_cell_table(n_cells=n_cells, n_markers=n_markers)
+ all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
+
+ all_clusters_format = pros.format_cell_table(cell_table=cell_table, clusters=all_clusters)
features = pros.featurize_cell_table(cell_table=all_clusters_format, featurization='cluster')
# at least 25 bootstrap iterations
with pytest.raises(ValueError, match="Number of bootstrap samples must be at least"):
@@ -127,10 +144,14 @@ def test_compute_topic_eda():
misc_utils.verify_in_list(eda_correct_keys=settings.EDA_KEYS, eda_actual_keys=list(eda.keys()))
-def test_create_difference_matrices():
+def test_create_difference_matrices(test_cell_table: Callable):
# Format & featurize cell table. Only test on clusters and 0.75 train frac to avoid repetition
- all_clusters = list(np.unique(TEST_CELL_TABLE[settings.CELL_TYPE]))
- all_clusters_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE, clusters=all_clusters)
+ n_markers = 10
+ n_cells = 1000
+ cell_table, all_markers = test_cell_table(n_cells=n_cells, n_markers=n_markers)
+ all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
+
+ all_clusters_format = pros.format_cell_table(cell_table=cell_table, clusters=all_clusters)
features = pros.featurize_cell_table(cell_table=all_clusters_format, featurization='cluster')
# create difference matrices
@@ -157,10 +178,12 @@ def test_create_difference_matrices():
assert diff_mat_infer['train_diff_mat'] is None
-def test_fov_density():
- # Format cell table
- all_clusters = list(np.unique(TEST_CELL_TABLE[settings.CELL_TYPE]))
- all_clusters_format = pros.format_cell_table(cell_table=TEST_CELL_TABLE, clusters=all_clusters)
+def test_fov_density(test_cell_table: Callable):
+ n_markers = 10
+ n_cells = 1000
+ cell_table, all_markers = test_cell_table(n_cells=n_cells, n_markers=n_markers)
+ all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
+ all_clusters_format = pros.format_cell_table(cell_table=cell_table, clusters=all_clusters)
cell_dens = pros.fov_density(all_clusters_format)
# check for correct names
diff --git a/tests/test_utils.py b/tests/test_utils.py
index 2fa228474..66cd52aa5 100644
--- a/tests/test_utils.py
+++ b/tests/test_utils.py
@@ -1,24 +1,33 @@
+from collections.abc import Mapping
from copy import deepcopy
+from functools import partial
from random import choices
from string import ascii_lowercase
-
+from anndata import AnnData
+from anndata.experimental import AnnCollection
import numpy as np
import pandas as pd
import synthetic_spatial_datagen
import xarray as xr
+from ark.utils.data_utils import AnnCollectionKwargs
import ark.settings as settings
-
-TEST_MARKERS = list('ABCDEFG')
+from typing import Tuple, List, Union
+try:
+ from typing import Unpack, Literal
+except ImportError:
+ from typing_extensions import Unpack
-def make_cell_table(num_cells, extra_cols=None):
- """ Generate a cell table with default column names for testing purposes.
+def make_cell_table(n_cells: int, n_markers: int, extra_cols: Mapping = None):
+ """Generate a cell table with default column names for testing purposes.
Args:
- num_cells (int):
+ n_cells (int):
Number of rows (cells) in the cell table
- extra_cols (dict):
+ n_markers (int):
+ Number of markers / channels in the cell table.
+ extra_cols (Mapping):
Extra columns to add in the format ``{'Column_Name' : data_1D, ...}``
Returns:
@@ -28,36 +37,63 @@ def make_cell_table(num_cells, extra_cols=None):
"""
# columns from regionprops extraction
- region_cols = [x for x in settings.REGIONPROPS_BASE if
- x not in ['label', 'area', 'centroid']] + settings.REGIONPROPS_SINGLE_COMP
- region_cols += settings.REGIONPROPS_MULTI_COMP
- # consistent ordering of column names
- column_names = [settings.FOV_ID,
- settings.PATIENT_ID,
- settings.CELL_LABEL,
- settings.CELL_TYPE,
- settings.CELL_SIZE] + TEST_MARKERS + region_cols + ['centroid-0', 'centroid-1']
+ region_cols = [
+ x for x in settings.REGIONPROPS_BASE if x not in ("label", "area", "centroid")
+ ]
+ post_marker_columns = [
+ settings.CELL_LABEL,
+ *region_cols,
+ *settings.REGIONPROPS_SINGLE_COMP,
+ *settings.REGIONPROPS_MULTI_COMP,
+ settings.FOV_ID,
+ settings.PATIENT_ID,
+ settings.CENTROID_0,
+ settings.CENTROID_1,
+ settings.CELL_TYPE,
+ ]
+
+ test_markers = [f"marker_{i}" for i in range(n_markers)]
+
+ cell_table_column_names = [settings.CELL_SIZE, *test_markers, *post_marker_columns]
if extra_cols is not None:
- column_names += list(extra_cols.values())
-
- # random filler data
- cell_data = pd.DataFrame(np.random.random(size=(num_cells, len(column_names))),
- columns=column_names)
- # not-so-random filler data
- centroids = pd.DataFrame(np.array([(x, y) for x in range(1024) for y in range(1024)]))
- centroid_loc = np.random.choice(range(1024 ** 2), size=num_cells, replace=False)
- fields = [(settings.FOV_ID, choices(range(1, 5), k=num_cells)),
- (settings.PATIENT_ID, choices(range(1, 10), k=num_cells)),
- (settings.CELL_LABEL, list(range(num_cells))),
- (settings.CELL_TYPE, choices(ascii_lowercase, k=num_cells)),
- (settings.CELL_SIZE, np.random.uniform(100, 300, size=num_cells)),
- (settings.CENTROID_0, np.array(centroids.iloc[centroid_loc, 0])),
- (settings.CENTROID_1, np.array(centroids.iloc[centroid_loc, 1]))
- ]
-
- for name, col in fields:
- cell_data[name] = col
+ cell_table_column_names += list(extra_cols.keys())
+
+ cell_data = pd.DataFrame(
+ data=np.empty((n_cells, len(cell_table_column_names))),
+ columns=cell_table_column_names,
+ )
+
+ rng = np.random.default_rng()
+
+ cell_data[settings.CELL_SIZE] = rng.integers(low=90, high=2000, size=n_cells)
+ cell_data[test_markers] = rng.random(size=(n_cells, n_markers))
+ cell_data[settings.CELL_LABEL] = np.arange(n_cells)
+
+ # Region Columns
+ for rc in region_cols:
+ cell_data[rc] = rng.random(size=n_cells)
+
+ # Region props single component
+ for rc in settings.REGIONPROPS_SINGLE_COMP:
+ cell_data[rc] = rng.random(size=n_cells)
+
+ # Region props multi component
+ for rc in settings.REGIONPROPS_MULTI_COMP:
+ cell_data[rc] = rng.random(size=n_cells)
+
+ # FOV ID
+ cell_data[settings.FOV_ID] = rng.integers(low=0, high=10, size=n_cells)
+
+ # Patient ID
+ cell_data[settings.PATIENT_ID] = rng.integers(low=0, high=5, size=n_cells)
+
+ # Centroid
+ cell_data[settings.CENTROID_0] = rng.integers(low=0, high=1024, size=n_cells)
+ cell_data[settings.CENTROID_1] = rng.integers(low=0, high=1024, size=n_cells)
+
+ # Cell Type
+ cell_data[settings.CELL_TYPE] = rng.choice(a=["A", "B", "C"], size=n_cells)
return cell_data
@@ -621,3 +657,135 @@ def generate_sample_fovs_list(fov_coords, fov_names):
)
return sample_fovs_list
+
+
+def generate_anndata_table(
+ rng: np.random.Generator,
+ n_obs: int,
+ n_vars: int,
+ fov_id: Union[str, int],
+ obs_properties: int,
+ obs_categorical_properties: int,
+) -> AnnData:
+ """Generates an AnnData Table with the following structure:
+
+ `AnnData`
+
+ ├── `X`: `n_obs` x `n_vars`
+
+ ├── `obs`: `n_obs` x (1 + `obs_properties` + `obs_categorical_properties`)
+
+ └── `obsm`: `n_obs` x 2
+
+ Args:
+ rng (np.random.Generator): The random number generator for reproducibility.
+ n_obs (int): The number of observations (cells, fiber segments, ezseg objects, etc...)
+ n_vars (int): The number of markers (channels).
+ fov_id (Union[str, int]): The FOV ID, can be a integer or a string used as a suffix.
+ `1` would make a FOV named `"fov_1"`, `"test"` would make a FOV named `"fov_test"`.
+ obs_properties (int): The number of floating point properties to add to the `obs` table.
+ obs_categorical_properties (int): The number of categorical properties to add to the
+ `obs` table.
+
+ Returns:
+ AnnData: The generated AnnData table.
+ """
+
+ _index = [f"cell_{i}" for i in range(n_obs)]
+
+ _X = pd.DataFrame(
+ data=rng.random(size=(n_obs, n_vars)),
+ index=_index,
+ columns=[f"channel_{i}" for i in range(n_vars)],
+ )
+
+ _obs = pd.DataFrame(
+ data={
+ settings.FOV_ID: [fov_id for _ in range(n_obs)],
+ settings.CELL_LABEL: np.arange(n_obs),
+ settings.CELL_TYPE: [
+ f"cell_type_{i}" for i in rng.integers(0, 10, size=n_obs)
+ ],
+ **{f"obs_prop_{i}": rng.random(size=n_obs) for i in range(obs_properties)},
+ **{
+ f"obs_cat_prop_{i}": [
+ f"obs_cat_prop_{j}" for j in rng.integers(0, 10, size=n_obs)
+ ]
+ for i in range(obs_categorical_properties)
+ },
+ },
+ index=_index,
+ )
+ _obsm = {
+ "spatial": pd.DataFrame(
+ data={
+ "centroid_y": rng.integers(0, 1024, size=n_obs),
+ "centroid_x": rng.integers(0, 1024, size=n_obs),
+ },
+ index=_index,
+ )
+ }
+
+ adata = AnnData(
+ X=_X,
+ obs=_obs,
+ obsm=_obsm,
+ )
+
+ return adata
+
+
+def generate_anncollection(
+ rng: np.random.Generator = np.random.default_rng(),
+ fovs: Union[int, list[str]] = 10,
+ n_obs: int = 100,
+ n_vars: int = 10,
+ obs_properties: int = 10,
+ obs_categorical_properties: int = 10,
+ random_n_obs: bool = True,
+ **anncollection_kwargs: Unpack[AnnCollectionKwargs],
+) -> Tuple[List[str], AnnCollection]:
+ """Generates an AnnCollection with the following parameters:
+
+
+ Args:
+ rng (np.random.Generator): The random number generator for reproducibility.
+ fovs (Union[int, list[str]]): The number of FOVs to generate, or a list of FOV IDs.
+ n_obs (int): The number of observations (cells, fiber segments, ezseg objects, etc...)
+ per FOV / AnnData Table.
+ n_vars (int): The number of markers (channels).
+ obs_properties (int): The number of floating point properties to add to the `obs` table.
+ obs_categorical_properties (int): The number of categorical properties to add to the
+ `obs` table.
+ random_n_obs (bool, optional): If True, `n_obs` is the upper bound for the number of
+ observations per FOV / AnnData Table.The number of `obs` is drawn from the discrete
+ uniform from [0, n_obs]. Defaults to True.
+
+ Returns:
+ Tuple[List[str], AnnCollection]: A list of FOV IDs and the generated AnnCollection.
+ """
+ if isinstance(fovs, int):
+ fovs = [f"fov_{i}" for i in range(fovs)]
+
+ adatas = {}
+
+ gen_ct_adata_func = partial(
+ generate_anndata_table,
+ rng=rng,
+ n_vars=n_vars,
+ obs_properties=obs_properties,
+ obs_categorical_properties=obs_categorical_properties,
+ )
+
+ for fov in fovs:
+ if random_n_obs:
+ n_obs_rand = rng.integers(n_obs)
+
+ fov_adata = gen_ct_adata_func(
+ n_obs=n_obs_rand, fov_id=fov
+ )
+ else:
+ fov_adata = gen_ct_adata_func(n_obs=n_obs, fov_id=fov)
+ adatas[fov] = fov_adata
+
+ return (fovs, AnnCollection(adatas=adatas, **anncollection_kwargs))
diff --git a/tests/utils/data_utils_test.py b/tests/utils/data_utils_test.py
index 384c9a0bf..053c87562 100644
--- a/tests/utils/data_utils_test.py
+++ b/tests/utils/data_utils_test.py
@@ -6,6 +6,7 @@
from shutil import rmtree
from typing import Generator, Iterator, List, Tuple
+import anndata.tests.helpers
import feather
import numpy as np
import pandas as pd
@@ -14,10 +15,13 @@
import xarray as xr
import numba as nb
import test_utils as ark_test_utils
+from anndata import read_zarr, AnnData
+from anndata.experimental import AnnCollection
+import dask.dataframe as dd
from alpineer import image_utils, io_utils, load_utils, test_utils
from ark import settings
from ark.utils import data_utils
-
+from typing import Callable
parametrize = pytest.mark.parametrize
@@ -77,7 +81,7 @@ def cell_table_cluster(rng: np.random.Generator) -> Generator[pd.DataFrame, None
Yields:
Generator[pd.DataFrame, None, None]: _description_
"""
- ct: pd.DataFrame = ark_test_utils.make_cell_table(num_cells=100)
+ ct: pd.DataFrame = ark_test_utils.make_cell_table(n_cells=100, n_markers=10)
ct[settings.FOV_ID] = rng.choice(["fov0", "fov1"], size=100)
ct["label"] = ct.groupby(by=settings.FOV_ID)["fov"].transform(
lambda x: np.arange(start=1, stop=len(x) + 1, dtype=int)
@@ -766,3 +770,139 @@ def test_stitch_images_by_shape(segmentation, clustering, subdir, stitching_fovs
# remove stitched_images from fov list
if not segmentation and not clustering:
stitching_fovs.pop()
+
+
+def test_convert_ct_fov_to_adata(tmp_path: pytest.TempPathFactory):
+ n_cells = 100
+ n_markers = 10
+ ct = ark_test_utils.make_cell_table(n_cells=n_cells, n_markers=n_markers)
+ ct_dd = dd.from_pandas(ct, npartitions=2)
+ fov1_dd = ct_dd[ct_dd[settings.FOV_ID] == 1]
+
+ var_names = [f"marker_{i}" for i in range(n_markers)]
+ obs_names = fov1_dd.drop(columns=var_names).columns.to_list()
+
+ fov1_adata_save_path = data_utils._convert_ct_fov_to_adata(
+ fov_dd=fov1_dd,
+ var_names=var_names,
+ obs_names=obs_names,
+ save_dir=tmp_path
+ )
+ save_path = fov1_adata_save_path.compute()
+
+ # Assert that the file exists
+ assert (tmp_path / "1.zarr").exists()
+
+ # Load the AnnData Zarr Store
+ fov1_adata = read_zarr(save_path)
+
+ # compute fov1_dd for asserts
+ fov1_df = fov1_dd.compute()
+
+ # Assert that the obs_names follow "{fov_id}_{cell_label}"
+ true_obs_names = list(map(lambda label: f"1_{int(label)}", fov1_df[settings.CELL_LABEL]))
+ assert fov1_adata.obs_names.tolist() == true_obs_names
+
+ # Assert that the X / Markers values are correct
+ np.testing.assert_allclose(actual=fov1_adata.X, desired=fov1_df[var_names].values)
+
+ # Assert that the obs columns are correct
+ expected_obs_columns = fov1_df.drop(
+ columns=[*var_names, settings.CENTROID_0, settings.CENTROID_1]
+ ).columns
+ assert fov1_adata.obs.columns.tolist() == expected_obs_columns.tolist()
+
+ # Assert that the obsm values are correct
+ np.testing.assert_allclose(
+ actual=fov1_adata.obsm["spatial"].values,
+ desired=fov1_df[[settings.CENTROID_0, settings.CENTROID_1]].values
+ )
+
+
+class TestConvertToAnnData:
+ @pytest.fixture(autouse=True)
+ def _setup(self, cell_table_cluster: pd.DataFrame, tmp_path_factory: pytest.TempPathFactory):
+ self.cell_table: pd.DataFrame = cell_table_cluster
+ self.ct_dir = tmp_path_factory.mktemp("cell_table")
+ self.cell_table_path = self.ct_dir / "cell_table.csv"
+ self.cell_table.to_csv(self.cell_table_path, index=False)
+
+ self.adata_dir = tmp_path_factory.mktemp("anndatas")
+
+ def test__init__(self):
+ cta = data_utils.ConvertToAnnData(self.cell_table_path)
+
+ assert set(cta.obs_names) == set(
+ self.cell_table.drop(
+ columns=[*[f"marker_{i}" for i in range(10)], settings.CELL_SIZE],
+ ).columns.to_list() + ["area"]
+ )
+ assert set(cta.var_names) == set([f"marker_{i}" for i in range(10)])
+
+ def test_convert_to_adata(self):
+ cta = data_utils.ConvertToAnnData(cell_table_path=self.cell_table_path,
+ markers="auto",
+ extra_obs_parameters=None)
+
+ adata_fov_paths = cta.convert_to_adata(self.adata_dir)
+
+ # Assert that the file exists
+ for fov, fov_adata_path in adata_fov_paths.items():
+ assert pathlib.Path(fov_adata_path).exists()
+
+
+@pytest.fixture(scope="module")
+def testing_anndatas(
+ tmp_path_factory: pytest.TempPathFactory
+) -> Callable[[int, pathlib.Path], Tuple[List[str], AnnCollection]]:
+ def create_adatas(n_fovs, save_dir: pathlib.Path):
+ fov_names, ann_collection = ark_test_utils.generate_anncollection(
+ fovs=n_fovs,
+ n_vars=10,
+ n_obs=100,
+ obs_properties=4,
+ obs_categorical_properties=2,
+ random_n_obs=True,
+ join_obs="inner",
+ join_obsm="inner"
+ )
+
+ for fov_name, fov_adata in zip(fov_names, ann_collection.adatas):
+ fov_adata.write_zarr(os.path.join(save_dir, f"{fov_name}.zarr"))
+ return fov_names, ann_collection
+
+ yield create_adatas
+
+
+def test_load_anndatas(testing_anndatas, tmp_path_factory):
+ ann_collection_path = tmp_path_factory.mktemp("anndatas")
+
+ fov_names, ann_collection = testing_anndatas(n_fovs=5, save_dir=ann_collection_path)
+
+ ac = data_utils.load_anndatas(ann_collection_path, join_obs="inner", join_obsm="inner")
+
+ assert isinstance(ac, AnnCollection)
+ assert len(ac.adatas) == len(fov_names)
+ assert set(ac.obs["fov"].unique()) == set(fov_names)
+
+ # Assert that each AnnData component of an AnnCollection is the same as the one on disk.
+ for fov_name, fov_adata in zip(fov_names, ann_collection.adatas):
+ anndata.tests.helpers.assert_adata_equal(
+ a=read_zarr(ann_collection_path / f"{fov_name}.zarr"),
+ b=fov_adata
+ )
+
+
+def test_AnnDataIterDataPipe(testing_anndatas, tmp_path_factory):
+ ann_collection_path = tmp_path_factory.mktemp("anndatas")
+
+ _ = testing_anndatas(n_fovs=5, save_dir=ann_collection_path)
+ ac = data_utils.load_anndatas(ann_collection_path, join_obs="inner", join_obsm="inner")
+
+ a_idp = data_utils.AnnDataIterDataPipe(fovs=ac)
+
+ from torchdata.datapipes.iter import IterDataPipe
+ assert isinstance(a_idp, IterDataPipe)
+
+ for fov in a_idp:
+ assert isinstance(fov, AnnData)
diff --git a/tests/utils/spatial_lda_utils_test.py b/tests/utils/spatial_lda_utils_test.py
index 86d8a5782..d33b91d0d 100644
--- a/tests/utils/spatial_lda_utils_test.py
+++ b/tests/utils/spatial_lda_utils_test.py
@@ -96,7 +96,7 @@ def test_check_featurize_cell_table_args():
def test_within_cluster_sums():
- cell_table = make_cell_table(num_cells=1000)
+ cell_table = make_cell_table(n_cells=1000, n_markers=10)
all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
formatted_table = pros.format_cell_table(cell_table, clusters=all_clusters)
featurized_table = pros.featurize_cell_table(formatted_table)
@@ -127,7 +127,7 @@ def test_plot_fovs_with_topics():
def test_save_spatial_lda_data():
- cell_table = make_cell_table(num_cells=1000)
+ cell_table = make_cell_table(n_cells=1000, n_markers=10)
all_clusters = list(np.unique(cell_table[settings.CELL_TYPE]))
cell_table_format = pros.format_cell_table(cell_table, clusters=all_clusters)
# test for non-existent directory
 +
+ +
+
+
+
+
+
 \n",
+ "
\n",
+ "