https://sciware.flatironinstitute.org/26_DataFormats
https://github.com/flatironinstitute/sciware/tree/main/26_DataFormats
Activities where participants all actively work to foster an environment which encourages participation across experience levels, coding language fluency, technology choices*, and scientific disciplines.
*though sometimes we try to expand your options
- Avoid discussions between a few people on a narrow topic
- Provide time for people who haven't spoken to speak/ask questions
- Provide time for experts to share wisdom and discuss
- Work together to make discussions accessible to novices
- Dedicated Zoom moderator to field questions.
- Please stay muted if not speaking. (Host may mute you.)
- We are recording. Link will be posted to https://sciware.flatironinstitute.org/
- Suggest topics or contribute to content in #sciware Slack
- We're recruiting: looking for CCB representative, contributors
- Intro to data and files
- Common file formats and their strengths/weaknesses
- Overview and python interfaces for HDF5 and ASDF
- Basic definitions
- Data-in-context
- Data's internal structure
- Particularly beyond lists/tables
- Desirable properties of file formats
- Overview of the trade-offs
This section is fairly abstract--don't worry, we'll be looking at examples soon!
Today is about the trade-offs imposed by different formats, and making choices that fit our situation and goals.
- Input to or output from a computational process
- Information we care about, such as:
- X, Y, Z component of a particle's position
- Edge weights on a graph
- Chunks of natural-language text
- ...
- Attempt to quantify some phenomenon
- A discrete entity on a filesystem
- A representation of some data
- using a finite series of bits
- on some persistent electronic storage
- separate from other data
- Organizing separate files is hard in its own right
- There are many ways to represent the same data
In memory!
- Representation trade-offs apply here too
- File-specific tasks include:
- Getting the right data to/from memory
- Persisting and duplicating data
- Data sharing/moving between systems
- Not the file extension (e.g.
.txt) - Particular choice of rules for:
- What data to store,
- How to represent it in bits
- How to organize it on disk
- Allowing disk-to-memory conversion
- in either direction
- consistently and repeatably
There are considerations outside our data that may influence our choice of file formats. Let's look at that.
- Some files are for publication
- These require the most care and robustness
- Some files are for internal use or short duration
- But listen to optimistic pessimism!
- It'll be more important & long-lived than you think
- Many fields have standard practice
- If you do something else, have a good reason
- "Data that describes data"
- Says how to interpret a representation/file
- For both the humans and the machines
- Might be explicit or implicit
- Expresses assumptions made about the data
- Data provenance? (Source, time, recording setup...)
- Hyperparameter choices that generated the data?
- Possible values for categorical observations
- e.g. 6 quark types, 4 DNA bases, ...
- "data dictionary"
-
In the file itself
- Most reliable & secure
- Takes more space
- Format must support it (somehow)
-
In the filename or directory structure
- Easy and straightforward
- until you need to share or re-sort your data
- leads to long filenames, incomplete capture
-
In a separate file
- Pro: Explicit, no special tooling needed
- Con: not automatic, can get lost
- Can be tricky if you have multiple writers
- In your head
- You know what's in
untitled_16_final_final.dat - I think those numbers mean...
- You know what's in
- In the paper
- No
- General tension: what is implicit vs explicit
Data often has some sort of natural internal structure. Here's how to describe that.
- Simple: only one observation or type of observation
- e.g. Height of an object at time
t=0,t=1, ... - One set of values, representing the same observable
- e.g. Height of an object at time
- Composite: more than one type of observation that go together as a unit
- e.g. Position & velocity of an object over time
- We mostly think about the composite case
- Fields are distinct observables
- the columns above, e.g.
mass,v_x, etc
- the columns above, e.g.
- Records are groups of fields that belong as a unit
- the rows above, i.e. one row per time
- A record is "one value in each of the fields"
- Does the row order mean anything?
- This can be a property of the data
- Mass and velocity mean different things
- Or it can be purely about representation
float32?float64? Ints? Strings?- Imperial? Metric? What's the zero value?
- For categorical data, what defines the category values?
- Are those values all known in advance?
- How do I encode those values?
- And your data are not lists/tables
- Those are representations of your data
- May or may not be a useful way to think about your data
- May or may not be a desirable file format
- First choice: what data to store?
- Limits of single-file table-oriented approach?
- We assumed that:
- each row/record represents a new time
- each time is equally far apart
- These are choices in representation
- i.e. choosing what not to represent
- Consider representing implicit fields explicitly
- All data is composite if you do
- Suppose your data is a 6d tensor
- What dimension (field) distinguishes the records?
- Is each record just one field with a 5d tensor?
- How would you slice along the different dimensions?
- Fields' interpretation might change
f_nn: but the nearest neighbor can vary
- Some fields don't apply to all records
- "Spouse" or "dependents" fields?
- What if we want to add a new field partway through?
- Partly a metadata issue, partly a data design issue
- Formats handle empty fields differently
- What if your table is mostly blank?
- Fields can evolve over different time scales or be treated at different spatial resolutions
- Mass gets repeated--can it even change?
- Adaptive grids, different-scale phenomena
- Natural solution: record them as different files
- How do I keep the files together?
- How do I link related records?
- Relational models address this (not in today's scope)
- Still useful for lots of scientific data
- Just be aware of the limitations
- It's expected that you'll find cases where data isn't neatly tabular
- Whatever problem you encounter, you probably aren't the first
- Ask around for advice!
Now that we've thought about data, let's look at how we would like to record it.
- This is Flatiron: we'll focus on efficiency and clarity
- There are plenty of other desirable properties for files
- Access control
- Resilience to data corruption
- Verified authenticity
- ...
- These are out of scope for today
- Read files fast
- Sequentially (from start to finish)
- Or skipping around (random access)
- Write files fast
- Use minimal disk space
- Open files on any machine
- Read files without special software
- Interpret the contents without outside knowledge
- Know when data is missing from a record
You can't have all of those things at once!
It's easy to come up with exceptions!
- Smaller files are faster to read than larger ones
- There's less data to transfer
- Flexibility and transparency require larger sizes
- Two-digit vs four-digit years
- Reading speeds compete with writing speeds
- "Time" may not be the best way to structure a file
- Random access competes with everything
- requires external indexes or regular-sized records
- but enables using a subset of the full file
- The extremes:
- Flexible, transparent, but big
- Concise, efficient, but brittle
- We sort of map these onto "human-readable" and "binary"
- But in practice all formats try to balance these extremes
- Best -- or worst -- of both worlds
Robert Blackwell (SCC)
- Text only
- A human can read, but not necessarily understand, it
- ...That's about it
- Self-documenting
- No special tooling required to inspect
- Typically easy to read, but maybe not understand
- Portable
- Generally flexible/extendable
$ cat Energy_K=1.0_Beta=2.5_Gamma=193.2
3.953190- EXTREMELY slow/space inefficient
- Difficult to extend
- Blows up very very quickly at scale
# t, x, v_x
0.00000, 0.00000, 0.15915
0.33333, 0.86603, -0.07958
0.66667, -0.86603, -0.07958
1.00000, -0.00000, 0.15915
- "Comma separated values"
- Fixed table
- Flexible types
- Comments
- Very portable
- Allows for empty fields
#t x v_x
0.00000 0.00000 0.15915
0.33333 0.86603 -0.07958
0.66667 -0.86603 -0.07958
1.00000 -0.00000 0.15915
- Often a homegrown extension of csv
- Space/tab/whatever delimited
- Non-standard, so might have to write custom parser
[{"t": 0.0, "x": 0.0, "v": 0.15915},
{"t": 0.33333, "x": 0.86603, "v": -0.07958},
{"t": 0.66667, "x": -0.86603, "v": -0.07958},
{"t": 1.0, "x": 0.0, "v": 0.15915}]- List of objects
- Handles missing fields
{"t": [0.0, 0.33333, 0.66666, 1.0],
"x": [0.0, 0.86603, -0.86603, 0.0],
"v": [0.15915, -0.07958, -0.07958, 0.15915]}- Object of lists
- Can't handle missing fields
- ...without special values
{'points': [{'x': [0.0, 0.0, 0.0], 'radius': 1.0},
{'x': [2.0, 0.0, 0.0], 'radius': 0.5}],
'params': {'dt': 0.0001,
't_final': 1.0,
'optimization': {'dt_opt': 0.1, 'shell': 1.5} },
'velocity_field': {'resolution': 0.5, 'dt_write': 0.5}}- Flexible/extendable -- very portable
- Verbose (con? pro? context matters.)
- No comments! (I assure you this is terrible)
jqutility useful for formatting
- Size: Typically 2-??X larger than machine native formats
- CSV/TXT: ~2x for floats
- JSON: minified ~2x, formatted ~10x (+?!)
- Performance: Lot of overhead to read - easy for humans, hard for computers
- Only sequential access -- No random access
- Sequential is a 'pro' for writing often
Lehman Garrison (SCC)
- Any file meant to be read by a machine instead of a human
- Low-level, often mostly a "raw memory dump"
- The values are stored identically on disk as they are in memory
- No expensive translation step, like with CSV to binary
- Or if there is a translation step (such as compression), it's very efficient
- Binary formats are almost always the right way to store large amounts of data!
Let's write the raw contents of an array with Numpy's tofile():
>>> a = np.arange(10**7, dtype='f4')
>>> a.tofile('mybinfile')$ head -c 128 mybinfile
�?@@@�@�@�@�@AA A0A@APA`ApA�A�A�A�A�A�A�A�A�A�A�A�A�A�A�A�A
- It's gobbledygook, as expected
- What would we do if we wanted it to be human readable?
Could make a human readable file with:
>>> np.savetxt('mytxtfile', a)$ head -n8 mytxtfile
0.000000000000000000e+00
1.000000000000000000e+00
2.000000000000000000e+00
3.000000000000000000e+00
4.000000000000000000e+00
5.000000000000000000e+00
6.000000000000000000e+00
7.000000000000000000e+00
- It's so much slower to save, so much slower to load, and so much bigger to store!
>>> %timeit np.savetxt("mytxtfile", a)
14.5 s ± 141 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
>>> %timeit a.tofile("mybinfile")
19.1 ms ± 196 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)$ ls -lh
-rw-rw-r-- 1 lgarrison lgarrison 39M Dec 8 12:09 mybinfile
-rw-rw-r-- 1 lgarrison lgarrison 239M Dec 8 12:09 mytxtfile
- Don't always use Numpy's
tofile()!- or similar raw binary dumps in other languages/packages
- Doesn't store metadata
- Not self-describing—how will a user know how to read? Does this file contain floats or ints? 4-byte or 8-byte? Etc.
>>> np.fromfile("mybinfile")
array([7.81250000e-03, 3.20000076e+01, 2.04800049e+03, ...,
5.88761292e+53, 5.88762023e+53, 5.88762753e+53])That's garbage! Have to remember to do:
>>> np.fromfile("mybinfile", dtype='f4')
array([0.000000e+00, 1.000000e+00, 2.000000e+00, ..., 9.999997e+06,
9.999998e+06, 9.999999e+06], dtype=float32)- Better formats exist that solve these problems
- Self-describing (e.g. knows its own
dtype) - Stores metadata (what simulation did this data come from?)
- Retain the performance and size benefits of raw binary
- Self-describing (e.g. knows its own
- Another key advantage of binary over human-readable is jump-ahead (next slide...)
- Example: I have a file with 1 billion elements, but I know I only need elements 500 million to 501 million
- In most human-readable formats, have to read all 501 million elements, then discard all the but last million
- In binary formats with fixed record size, you can compute:
- Each record (array element) is (e.g.) three 4-byte floats: 12 bytes per record
- 500 millionth record is at offset: 12*500M = 6 billion bytes
- Start reading from the 6 billionth byte in the file, save tons of time
- A good binary format (HDF5, ASDF) will handle this for you under the hood
There are thousands of binary formats! Here are a few common, high-quality ones:
- HDF5
- ASDF
- Numpy .npy
- Msgpack
- Python Pickle
- Other domain-specific formats
- HDF5
- Wide adoption, feature-rich, often the Right Choice
- Will talk about with examples in Part 2
- Good for: arrays of data with metadata, complex data models, inter-language operability
- Downsides: complicated API, opaque internals (unpredictable performance)
- https://docs.h5py.org/en/stable/quick.html#core-concepts
- ASDF
- Combines human-readable header with binary data blocks
- Good for: exchanging data (small or big) between Python users, medium-complicated use-cases like compression, extendable arrays, etc.
- Downsides: limited support outside Python
- https://asdf.readthedocs.io/en/stable/asdf/overview.html
- .npy/.npz files (Numpy specific)
- Simple format for Numpy arrays; use with
np.save()andnp.load() - .npz is literally zip file for .npy files
- Good for: data consists only of Numpy arrays, easy to read in other languages
- Downsides: only works with Numpy arrays, no metadata support besides types
- https://numpy.org/doc/stable/reference/generated/numpy.save.html
- Simple format for Numpy arrays; use with
- Msgpack
- "Binary JSON": lists, key-value dicts, etc
- Good for: arbitrarily structured binary data
- Downsides: bad for arrays: no random access/jump-ahead (each item can have different size)
- https://msgpack.org/index.html
- Python Pickle
- Stores arbitrary Python objects
- Good for: saving and loading intermediate state of custom Python objects
- Downsides: slow, fragile (won't work across Python versions or if packages are missing)
- https://docs.python.org/3/library/pickle.html
- Domain-specific formats
- Countless domain-specific formats!
- e.g. NIfTI (neuroscience), netCDF (geospatial)
- Good for: compatibility with software tools in a domain
- Downsides: users outside that domain won't know what to do with the file
- Binary formats store data in a low-level way, often as a direct copy of the values in memory
- Nearly always the right choice when size or performance is a consideration
- Downsides: the format you choose locks you (and your users) into that format's tooling ecosystem
- HDF5 files have to be read with an HDF5 library, npy files have to be read with numpy, etc.
- Usually can't open a file and know what's inside without that format's tools (but see ASDF)
- Other formats you've used? Things you like? Challenges?
- Questions?
Please give us some feedback!
https://bit.ly/sciware-file-formats-2022
Dylan Simon (SCC)
Largely taken from HDFGroup's Introduction to HDF5
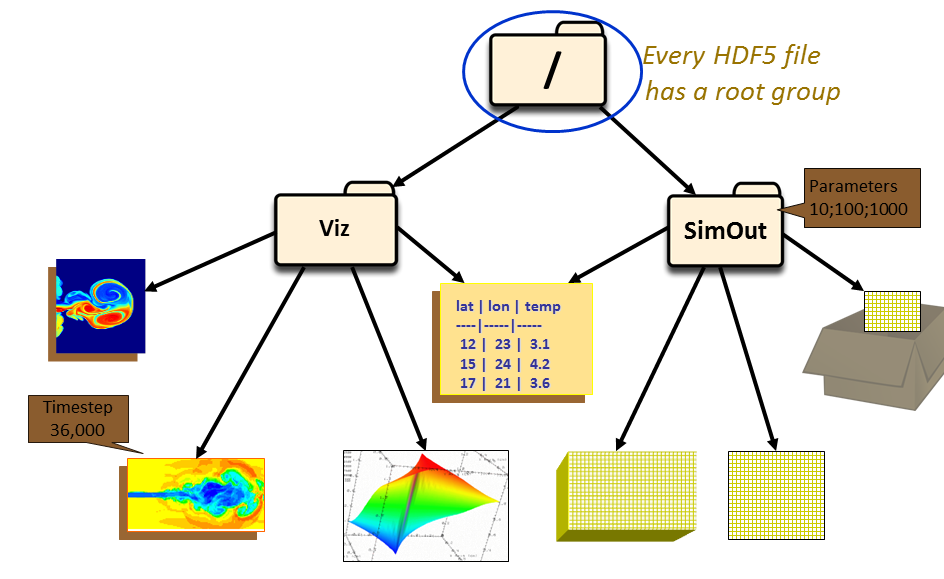
"An HDF5 file can be thought of as a container (or group) that holds a variety of heterogeneous data objects (or datasets). The datasets can be images, tables, graphs, and even documents, such as PDF or Excel"
Groups are like directories in a filesystem
- UNIX path names are used to reference objects:
/SimOut/Mass
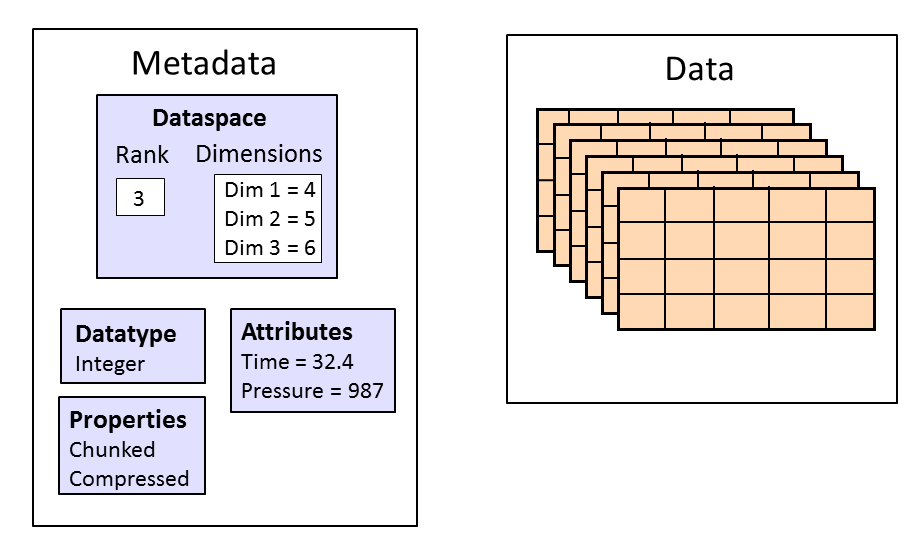
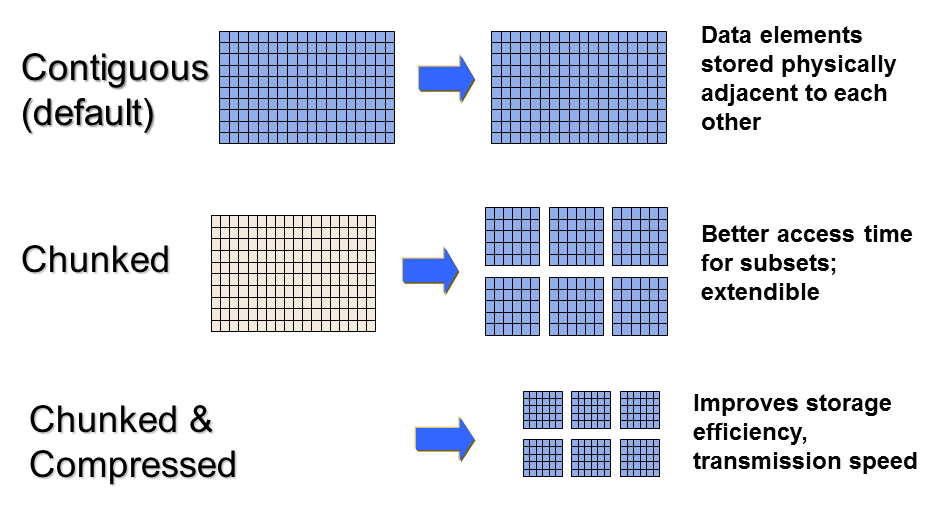
- Datasets store multi-dimensional blocks of data of a single type
- Usually represent a single "field" of data
- Datatypes are designed to map to in-memory representations
- Predefined datatypes: atomic values
H5T_IEEE_F32LE,H5T_NATIVE_FLOAT(float)H5T_INTEL_I32,H5T_STD_I32LE,H5T_NATIVE_INT(int)H5T_C_S1(char, basis for fixed-length strings)
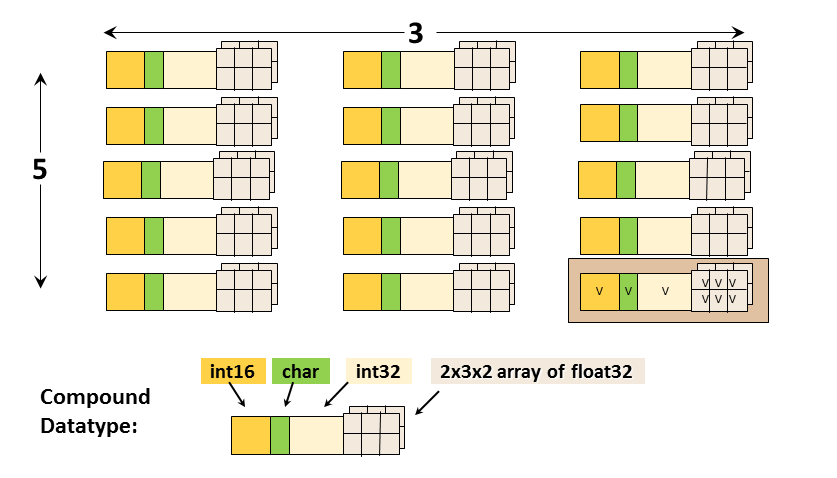
- Derived datatypes: compound values composed of other types
- fixed-length strings for random access
- Compound types can be nested and include fixed-sized arrays
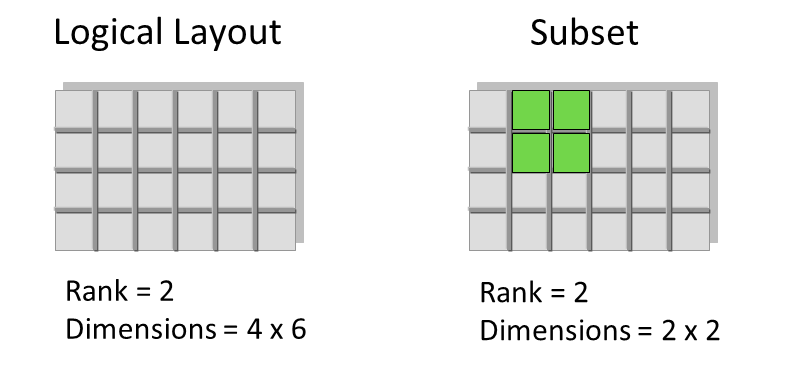
- Dataspaces represent the dimensions of an array (scalar, vector, etc.)
- Can be used to describe overall Dataset size, and subsets ("views", "slices") of data
- Attributes are metadata attached to an object (usually a Group or Dataset)
- Have names and values
- Values are the same as Dataset values, but are usually small
- Scalar value of simulation parameter
- String describing field, units, etc.
- Properties control how data is stored on disk
- Links (symlinks), Filters (compression, checksums)
- Images, tables, units
- Default values (missing data can be hard in binary)
- h5ls
- h5dump
- hdfview
- Like ls for hdf5 structure
- Useful for summarizing contents
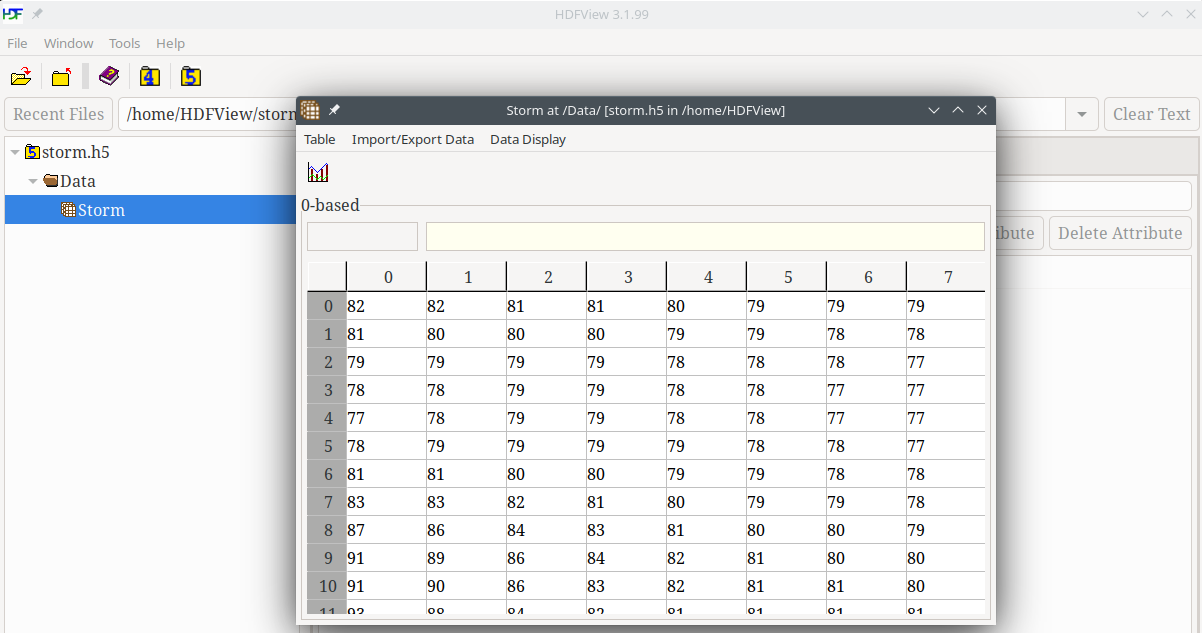
h5ls -r
/ Group
/results Group
/results/count Dataset {SCALAR}
/results/final Group
/results/final/x Dataset {1000}
/results/final/y Dataset {1000}
/results/final/Velocity Dataset {1000, 2}
/results/final/Mass Dataset {1000}
- Full text dump of all data
- Can limit to certain groups (
-g /path) or dataset (-d /path/x)
HDF5 "vasp.h5" {
GROUP "/" {
GROUP "/results" {
DATASET "count" {
DATATYPE H5T_STD_I64LE
DATASPACE SCALAR
DATA {
(0): 20
}
}
GROUP "final" {
DATASET "x" {
DATATYPE H5T_IEEE_F64LE
DATASPACE SIMPLE { ( 1000 / 1000 ) }
DATA {
(0): 0.052104
(1): 0.139832
(2): 1.398349
...
}
ATTRIBUTE "units" {
DATATYPE H5T_STRING {
STRSIZE 8;
STRPAD H5T_STR_NULLPAD;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "furlongs"
}
}
}
}
}
}
- Graphical viewer (and editor) for hdf5 files
- There is only one full HDF5 implementation (in C): hdf5
- Many interfaces built on top of this
/* Open an existing file. */
hid_t file_id = H5Fopen(FILE, H5F_ACC_RDWR, H5P_DEFAULT);
/* Open an existing dataset. */
hid_t dataset_id = H5Dopen2(file_id, "/dset", H5P_DEFAULT);
/* Read the dataset. */
int dset_data[4][6];
herr_t status = H5Dread(dataset_id, H5T_NATIVE_INT, H5S_ALL, H5S_ALL, H5P_DEFAULT, dset_data);
/* Close the dataset. */
status = H5Dclose(dataset_id);
/* Close the file. */
status = H5Fclose(file_id);- h5py is the standard interface to hdf5 in python
- Provides a low-level interface (that looks a lot like the C API)
- More common high-level interface provides pythonic access
- Open files with
h5py.File("file.h5", mode)modecan be"r"(read)"w"(create/truncate), or"r+"(read/write)
- Files can be closed using
with(explicit scope) or automatically when unreferenced - Groups provide dict-like interface, index with
[]and paths
file = h5py.File("file.h5", "r")
# <HDF5 file "file.h5" (mode r)>
group = file["results"]["final"]
group = file["results/final"]
# <HDF5 group "/results/final" (4 members)>- Datasets mostly look like numpy arrays, can be indexed with
[] - Scalar values are accessed by
()
count = file["results/count"]
# <HDF5 dataset "count": shape (), type "<i8">
count[()]
# 20
x = group["x"]
# <HDF5 dataset "x": shape (1000), type "<f8">
x.dtype
x.shape
x[0]
# 0.052104: numpy.float64
x[10:20]
# numpy.ndarray- Groups and Datasets have
attrs - Works like a simple dictionary
group.attrs.keys()
x.attrs["units"]with h5py.File("file.h5", "w") as file:
group = file.create_group("results/data")
x = group.create_dataset("x", (1000,), dtype="f")
x[0] = 0.052104
y = group.create_dataset("y", data=np.arange(1000))
x.attrs["units"] = "furlongs"pos = group.create_dataset("Velocity", (1000,2), dtype="f")
pos[10,1] = 3.5
pos[10][1] # works for access, not assignment!
pos[0,:]
pos[20:29,0] = numpy.arange(10)- HDF5 allows incremental resizing of Datasets (like appending to a CSV)
mass = f.create_dataset("Mass", (10,), maxshape=(None,))
mass[0:9] = ...
mass.resize((20,))
mass[10:19] = ...- Avoid opening files (or objects) repeatedly
- Combine related data in the same file (and...)
- Add any relevant metadata in attributes
- software version, timestamp, parameters, configuration
- parameter descriptions, units
- HDF5 provides a parallel-enabled (MPI) interface: you don't generally need it
- better to do all writes from a single rank
- https://github.com/TRIQS/h5: C++, python
Lehman Garrison
- Most binary formats are all binary—no way to know what's inside with installing special tools (e.g.
h5ls) - Doesn't have to be that way, we can have a human-readable header while still storing the data in binary blocks for efficiency
- That's what ASDF is!
- ASDF = Advanced Scientific Data Format
- "A next-generation interchange format for scientific data"
- https://github.com/asdf-format/asdf
import asdf
import numpy as np
seed = 123
rng = np.random.default_rng(seed)
sequence = np.arange(10**6, dtype='f4')
rand = rng.random((512,512))
tree = {'seq': sequence,
'rand': rand,
'params':
{'comment': 'This is an example for Sciware',
'seed': seed,
},
}
af = asdf.AsdfFile(tree)
af.write_to('/tmp/myfile.asdf')#ASDF 1.0.0
#ASDF_STANDARD 1.5.0
%YAML 1.1
%TAG ! tag:stsci.edu:asdf/
--- !core/asdf-1.1.0
asdf_library: !core/software-1.0.0 {author: The ASDF Developers, homepage: 'http://github.com/asdf-format/asdf',
name: asdf, version: 2.13.0}
history:
extensions:
- !core/extension_metadata-1.0.0
extension_class: asdf.extension.BuiltinExtension
software: !core/software-1.0.0 {name: asdf, version: 2.13.0}
params: {comment: This is an example for Sciware, seed: 123}
rand: !core/ndarray-1.0.0
source: 1
datatype: float64
byteorder: little
shape: [512, 512]
seq: !core/ndarray-1.0.0
source: 0
datatype: float32
byteorder: little
shape: [1000000]
...
<D3>BLK^@0^@^@^@^@^@^@^@^@^@^@^@^@^@= ^@^@^@^@^@^@= ^@^@^@^@^@^@= ^@<B7>_<98><E3>^B<87><A5><FE>^M
^M<BF><A7>^H*<B0><91>*^@^@^@^@^@^@<80>?^@^@^@@^@^@@@^@^@<80>@^@^@<A0>@^@^@<C0>@^@^@<E0>@^@^@^@A^@^@^PA^@import asdf
sequence = list(range(12))
squares = {k:k**2 for k in range(10,20)}
tree = { 'seq': sequence, 'squares': squares}
af = asdf.AsdfFile(tree)
af.write_to('/tmp/myfile.asdf')#ASDF 1.0.0
#ASDF_STANDARD 1.5.0
%YAML 1.1
%TAG ! tag:stsci.edu:asdf/
--- !core/asdf-1.1.0
asdf_library: !core/software-1.0.0 {author: The ASDF Developers, homepage: 'http://github.com/asdf-format/asdf',
name: asdf, version: 2.13.0}
history:
extensions:
- !core/extension_metadata-1.0.0
extension_class: asdf.extension.BuiltinExtension
software: !core/software-1.0.0 {name: asdf, version: 2.13.0}
seq: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11]
squares: {10: 100, 11: 121, 12: 144, 13: 169, 14: 196, 15: 225, 16: 256, 17: 289,
18: 324, 19: 361}- Development led by astronomy community (STScI) as a FITS replacement (all JWST data in ASDF)
- Greenfield, et al. (2015) lays out motivation: want human-readable headers; avoid complexity of HDF5 (is it really archival?); HDF5 doesn't represent certain data structures (e.g. Python dict) well
- The reference implementation is in Python. Excellent integration with Numpy. C++ interface less mature.
- Lehman prefers to use ASDF over HDF5 when it makes sense
- End-users are using Python and are okay with non-HDF5
- Reasonably feature-complete
- Multiple compression options
- Streaming blocks (for incoming data of unknown size)
- Lazy-loading of arrays
- Memory-mapped IO for efficient jump-ahead
- Can define extensions and schema for serializing custom types
- File format is formally standardized
Please give us some feedback!