-
Notifications
You must be signed in to change notification settings - Fork 58
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
MCMICRO compatibility with in situ sequencing #502
Comments
|
Maybe I should specify the |
|
How many input tiff files do you have and what are the image dimensions? Do you have one whole-slide image for each mRNA transcript, or do you have many smaller "tiles" for each transcript? |
|
|
I wanted to follow up on this thread since I was wondering whether mcmicro will support a module for ISS decoding across multiple rounds, or whether that is still in the pipeline? Thank you! |
|
What software do folks currently use for ISS decoding? If there is an existing containerized module we can leverage, it would be relatively straightforward to add one to the pipeline. |
|
I guess spacetx/starfish offers a lot of flexibility (in terms of different decoding schemes) - https://spacetx-starfish.readthedocs.io/en/latest/user_guide/index.html#spot-based-decoding. Additionally, I must admit I have not been able to figure out how to implement spot detection with 3seg yet - I`ve been trying a few params.yml modifications I found https://mcmicro.org/troubleshooting/tuning/s3seg.html, however, I keep getting an error that the puncta parameters are not supported. Is there an optimum configuration for the params.yml required for spot detection to work? Thanks ever so much for your help! |
|
Hi @Boehmin Can you list the parameters you are using for s3seg spot detection please? Thanks. |
|
I added the following to my s3seg options: |
|
Hi @Boehmin , thanks for listing your s3seg parameters. Your spot detection parameters look valid. However, it appears you're using the newer version of s3seg and unfortunately, we have not had a chance to port the spot detection capabilities that were in the older version yet, which is why it is erroring. |
|
Hi @clarenceyapp , thanks a mill for your help - that`s great news! I use s3seg version: 1.5.4-large if that helps as well. Which version do you recommend I go for? The image is 21,869x17,528 pixels so should be ok then? |
|
Another thing to note is that s3seg would only detect the x-y coordinates, so additional processing would need to happen for trace building and spot detection. It seems that would work. |
Hi @jmuhlich, thanks to you and your team for providing this easy-to-use software to the community! I'd like to try MCMICRO on my ISS (in situ sequencing) data, which belongs to a branch in spatial transcriptomics.
What I have done
exemplar-001data, and the output folder structure is like:The data structure of ISS
Briefly, ISS detects mRNA on the whole tissue slides using cyclic imaging and barcode decoding.
Input1: raw image files (in .tif format)
The raw images obtained from an ISS experiment are almost the same as that of
exemplar-001, but only differ in the:.tifformat, rather than.ome.tif.Input2: markers
This is the screenshot of my
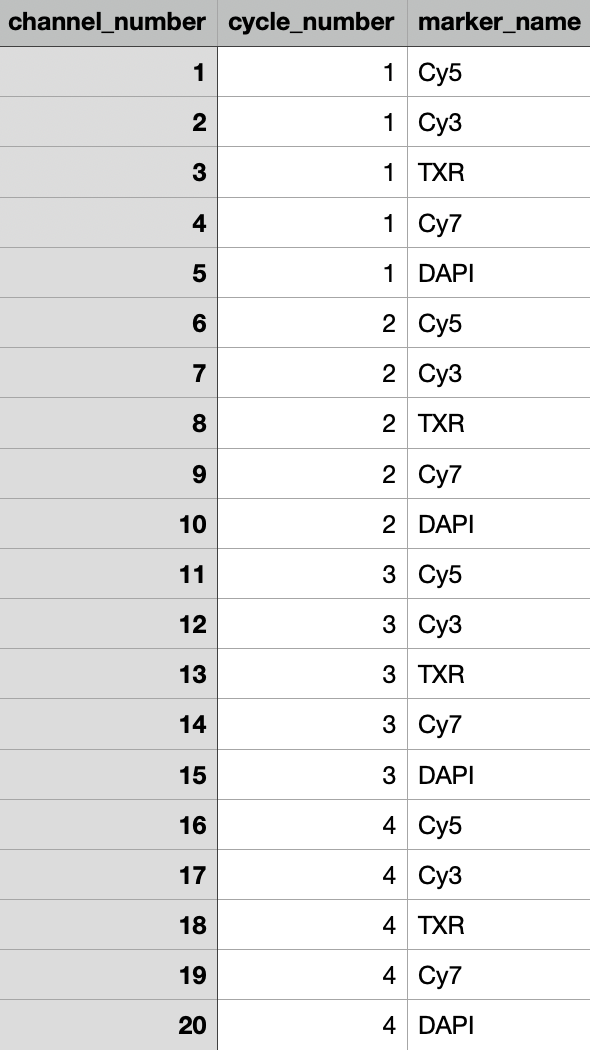
markers.csv.Question
I've tested MCMICRO on my ISS data, however, it failed on
quantification:mcquantstep which alerted,Exception: The number of channels in markers.csv doesn't match the image. Stitching and alignment can run quickly, despite the output was incorrect after visual inspection.Thanks for your patience!
The text was updated successfully, but these errors were encountered: