+Testing Containers and WebAssembly in Submissions to the FDA +
+ +-Testing Containers and WebAssembly in Submissions to the FDA -
- -The R Consortium Submission Working Group has now successfully made two pilot submissions to the FDA. All the submissions done by the group are focused on improving practices for R-based clinical trial regulatory submissions. Now, the R submission Working Groups in collaboration with Appsilon and Posit are exploring new technologies such as Containers and WebAssembly. In this article, we dive into the details of this exploration.
+How Everything Started
+Pilot 1
+This pilot was initially submitted on November 22, 2021. This submission was the first publicly available R-based submission to the FDA. This was a test submission that aimed to explore the submission of an R package to the FDA following the eCTD specifications. The submission included an R package, R scripts for analysis, R-based analysis data reviewed guide (ADRG), and other important components. The final response letter from the FDA was received on March 14, 2022.
+Pilot 2
+This was one of the first submission packages containing a Shiny application. The main goal of this pilot was to test the submission of an R-based Shiny application bundled into a submission package and transfer it successfully to FDA reviewers. The submitted application was built using the datasets and analysis that were used for the R Submission Pilot 1. The deployed version of this application is available on this site. Alternatively, a Rhino-based version of the application can be found here.
+The final response letter from the FDA was reviewed on September 27, 2023.
+
In this submission, there were many open-source R packages that were used to create and execute the Shiny application. A very well known shiny-based interactive exploration framework {teal} was used mainly for analyzing the clinical trial data; this package is included in the pharmaverse package repository. The full list of open-source and proprietary R analysis packages is available on this Analysis Data Reviewer’s Guide prepared by the R Consortium R Submissions Working Group for the Pilot 2.
+What’s next?
+Pilot 3
+This pilot was successfully submitted to FDA on Aug 28, 2023. This was the first publicly available R submission that included R scripts to produce ADaM datasets and TLFs. Both the ADaMs (SDTM .xpt sources from the CDISC Pilot study) and the TLFs (ADaMs .xpt sourced from the ADaMs generated in R by the Pilot 3 team) were created using R. The next steps for this pilot is to await FDA’s review and approval which may take several months to complete.
+Pilot 4
+This pilot aims to explore using technologies such as containers and WebAssembly software to package a Shiny application into a self-contained unit, streamlining the transfer and execution process for enhanced efficiency.
+This pilot is expected to be divided into two parallel submissions:
+(a) will investigate WebAssembly and
+(b) will investigate containers.
+The Journey with WebAssembly and Containers
+Our team at Appsilon teamed up with the dynamic Pilot 4 crew to explore WebAssembly technology and containers. George Stagg and Winston Chang also joined the working group to discuss the web-assembly portion of Pilot 4. This partnership brought together our engineering prowess to contribute to these tools, injecting fresh perspectives into the ongoing pilot project.
+Some of the outcomes of the collaboration:
+-
+
We were able to set up a robust container environment for this pilot project.
+We aided the progress made on the use of both experimental technologies: containers and WebAssembly.
+We developed a working prototype submission using Podman container technology.
+We developed a working early-stage prototype for wrapping a small Shiny application using WebAssembly.
+
WebAssembly
+
WebAssembly allows languages like R to be executed at near-native speed directly within web browsers, providing users with the ability to run R code without having R installed locally. WebR is essentially the R programming language adapted to run in a web browser environment using WebAssembly. This project is under active development.
+The Pilot 4 Shiny App Up and Running on webR!
+
The deployed example of the Shiny app running on webR is available here. Check out the video of the application running below.
+ +During this pilot, engineers at Appsilon developed a prototype of a Shiny application running on webR. The application reuses most of the code from the previous pilot apps with some tweaks and a couple of hacks/changes to get around non CRAN dependencies, specially for data loading, WebR compatibilities, and shimming some of the functionality from {teal} and other packages that are (for now) not available on CRAN.
+webR Shiny App
+During the second iteration, which was recently held, Pedro Silva shared the process of developing this Shiny app running on webR.
+The Process
+-
+
Leverage last 2 iterations of the application
+-
+
Reuse as much code as possible
+Avoid touching the logic part
+
+Restrict the number of dependencies to packages on CRAN
+-
+
- Replace/shim functionality that was lost from removing dependencies +
+
Here is the list of dependencies to packages on CRAN, those that worked are colored green, those that were removed are marked in orange. We ended up with just 3 problematic dependencies (bold).
+| | |
+|------------------------------------|------------------------------------|
+| [library(config)]{style="color:orange"} | [library(reactable)]{style="color:green"} |
+| **library(cowplot)** | [library(rhino)]{style="color:orange"} |
+| [library(dplyr)]{style="color:green"} | [library(rtables)]{style="color:green"} |
+| [library(emmeans)]{style="color:green"} | [library(shiny)]{style="color:green"} |
+| [library(ggplot2)]{style="color:green"} | [library(stringr)]{style="color:green"} |
+| [library(glue)]{style="color:green"} | **library(teal)** |
+| [library(haven)]{style="color:green"} | **library(teal.data)** |
+| [library(htmltools)]{style="color:green"} | [library(tibble)]{style="color:green"} |
+| [library(huxtable)]{style="color:green"} | [library(tidyr)]{style="color:green"} |
+| [library(magrittr)]{style="color:green"} | [library(tippy)]{style="color:green"} |
+| [library(markdown)]{style="color:green"} | [library(Tplyr)]{style="color:green"} |
+| [library(pkgload)]{style="color:green"} | [library(utils)]{style="color:green"} |
+| [library(purrr)]{style="color:green"} | [library(visR)]{style="color:green"} |Issues with library(cowplot):
+-
+
- Some issue with low-level dependencies when deployed +
Solution:
+-
+
- Replace functionality with HTML +
Issues with library(teal):
+-
+
- Uses {shiny.widgets} (not working for webR) +
Solution:
+-
+
Redo the UI
+Load modules directly
+Recreate filter functionality
+
Issues with library(teal.data):
+-
+
- Use rds exports +
Solution:
+-
+
- Shim functionality, load data directly +
-
+
Leverage shinylive and httpuv to export and serve the application
+-
+
Shinylive can help streamline the export process
+-
+
Problems
+-
+
shiny.live won’t let us have non-R files in the application directory - this is an outstanding bug that George asked us to raise an issue for.
+We wouldn’t be able to run the application as a traditional shiny app
+
+Solution:
+
1. **Custom build script**
+{httpuv} can help serve the application
+-
+
- {httpuv} would run natively on a machine to serve the Shiny app +
+
+
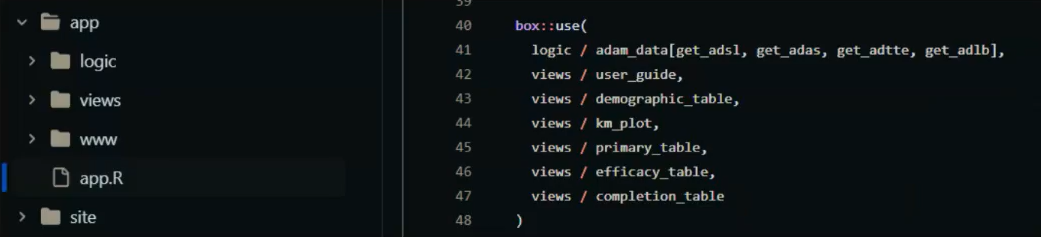
Application Structure
+The figure below shows an overview of what we ended with:
+
Some of the issues and solutions found at the very beginning:
+-
+
- The previous applications were built using golem and another one in Rhino; the support for these frameworks is not great in webR up to now.
+
-
+
- Solution
+
-
+
- {box} works out of the box (reuse the rhino version modules) +
- Simplify the structure and use a simple shiny modular structure +
+
+ - Solution
+
- Shinylive does not like non-R files when generating the bundle
+
-
+
- Solution
+
-
+
- Keep the app folder as clean as possible for now (www folder only) +
+
+ - Solution
+
- {teal} and {teal.data} are not on CRAN
+
-
+
- Solution
+
-
+
- Shim and used functionality +
- Use a simple tab system for the UI structure +
+
+ - Solution
+
The FDA were previously told that the shiny application being prepared for the Pilot 4 submission would not be a 1 to 1 mapping from the previous one submitted for the Pilot 2 due to certain constraints such as {teal} not being on CRAN; however, this didn’t represent a problem for them since they would mainly like to test the technology.
+Pedro Silva, one of the engineers working on the development of this app mentioned “While WebR is still in development, it shows tremendous promise! The loading is definitely still a pain point (over 100mb to set up the environment!) but it will only get better moving forward.”
+Containers
+
Containerization, particularly through technologies like Docker, Podman or Singularity, offers several advantages for deploying Shiny apps.
+Choosing the Right Container
+Choosing the right container was a question that arose in this project. Although Docker is the most popular, we decided to move forward with Podman.
+In our exploration of containerization tools for deploying Shiny applications, we’ve identified key distinctions between Docker and Podman that influenced our choice.
+Podman stands out for its daemonless architecture, enhancing security by eliminating the need for a central daemon process. Unlike Docker, Podman supports running containers as non-root users, a critical feature for meeting FDA reviewer requirements. Developed by Red Hat and maintained as an open-source project, Podman prioritizes security with its rootless container support, offering a robust solution for security-conscious users.
+Goals
+A Container-based method to deploy Pilot 2 Shiny App.
+What we did
+-
+
Configurable Podman Dockerfile / docker-compose.yml
+-
+
- R version +
- Registry / organization name / image name (differences between docker.io and ghcr.io) +
+Documentation on creating the container
+CI: Automated build on amd64 and arm64 platforms
+
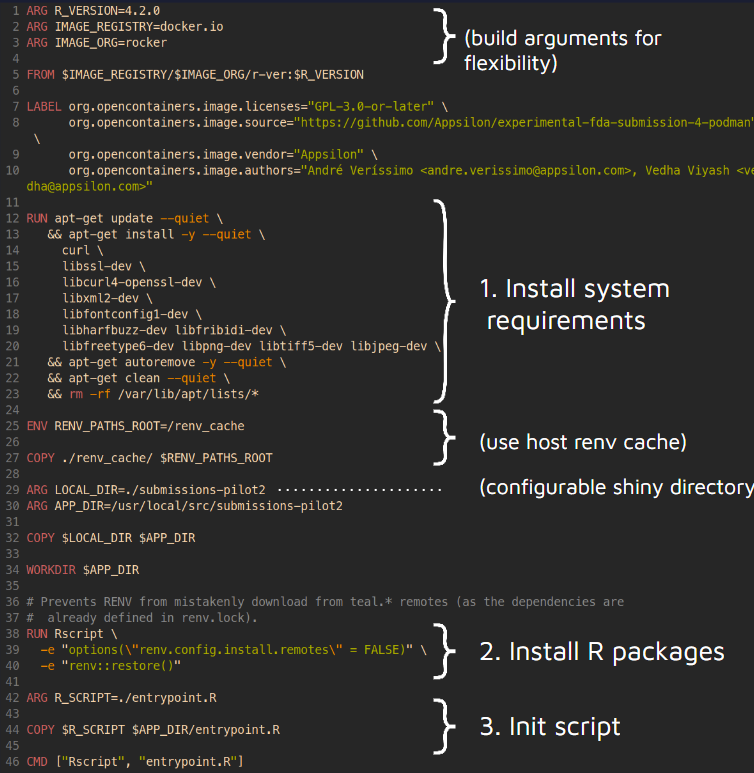
Podman short-demo
+ +Below is the dockerfile (recipe) for the container:
+
Next Steps
+The next steps are waiting for the review of the Pilot 3 by the FDA and to submit the two sections that explore the new technologies to regulatory authorities. Thanks to the collaboration between the R Submission Working Group and other institutions, there is already a working prototype of a {teal}-like Shiny application running on webR and further exploration with Podman is underway.
+-
+
- Submission to FDA +
- Rhino Compatibility +

Appsilon is working on the side with Rhino compatibility, eventually we might be able to just add this framework into the Pilot 4 application.
+-
+
{teal}
+This and other packages might be on CRAN soon. We could incorporate them after that, replacing the shims created for this version
+Boot Time
+We need to improve boot time (remove dependencies and keep working on webR).
+
Reuse
Citation
@online{veríssimo2024,
+ author = {Veríssimo, André and Makowski, Tymoteusz and Silva, Pedro
+ and Viyash, Vedha and , APPSILON},
+ title = {Testing {Containers} and {WebAssembly} in {Submissions} to
+ the {FDA}},
+ date = {2024-02-01},
+ url = {https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html},
+ langid = {en}
+}
+Last updated
2024-02-01 12:24:29.879149
+2024-02-01 12:42:36.670717
Details
Last updated
2024-02-01 12:24:37.177593
+2024-02-01 12:42:44.050491
Details
Last updated
2024-02-01 12:24:38.870695
+2024-02-01 12:42:45.713213
Details
Last updated
2024-02-01 12:24:06.874564
+2024-02-01 12:42:12.894734
Details
As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
Last updated
2024-02-01 12:24:43.976417
+2024-02-01 12:42:49.540771
Details
Last updated
2024-02-01 12:24:41.550192
+2024-02-01 12:42:47.195254
Details
Last updated
2024-02-01 12:24:26.757735
+2024-02-01 12:42:33.536958
Details
Last updated
2024-02-01 12:24:20.344918
+2024-02-01 12:42:27.242831
Details
Last updated
2024-02-01 12:24:33.442683
+2024-02-01 12:42:40.324505
Details
Last updated
2024-02-01 12:24:18.44166
+2024-02-01 12:42:25.336389
Details
Last updated
2024-02-01 12:24:09.63242
+2024-02-01 12:42:16.798817
Details
Last updated
2024-02-01 12:24:34.960458
+2024-02-01 12:42:41.841083
Details
Last updated
2024-02-01 12:24:31.481975
+2024-02-01 12:42:38.30988
Details
Last updated
2024-02-01 12:24:14.841985
+2024-02-01 12:42:21.757191
Details
Last updated
2024-02-01 12:24:16.723448
+2024-02-01 12:42:23.618603
Details
Last updated
2024-02-01 12:24:22.445948
+2024-02-01 12:42:29.277076
Details
The R Consortium Submission Working Group has now successfully made two pilot submissions to the FDA. All the submissions done by the group are focused on improving practices for R-based clinical trial regulatory submissions. Now, the R submission Working Groups in collaboration with Appsilon and Posit are exploring new technologies such as Containers and WebAssembly. In this article, we dive into the details of this exploration.
-How Everything Started
-Pilot 1
-This pilot was initially submitted on November 22, 2021. This submission was the first publicly available R-based submission to the FDA. This was a test submission that aimed to explore the submission of an R package to the FDA following the eCTD specifications. The submission included an R package, R scripts for analysis, R-based analysis data reviewed guide (ADRG), and other important components. The final response letter from the FDA was received on March 14, 2022.
-Pilot 2
-This was one of the first submission packages containing a Shiny application. The main goal of this pilot was to test the submission of an R-based Shiny application bundled into a submission package and transfer it successfully to FDA reviewers. The submitted application was built using the datasets and analysis that were used for the R Submission Pilot 1. The deployed version of this application is available on this site. Alternatively, a Rhino-based version of the application can be found here.
-The final response letter from the FDA was reviewed on September 27, 2023.
-
In this submission, there were many open-source R packages that were used to create and execute the Shiny application. A very well known shiny-based interactive exploration framework {teal} was used mainly for analyzing the clinical trial data; this package is included in the pharmaverse package repository. The full list of open-source and proprietary R analysis packages is available on this Analysis Data Reviewer’s Guide prepared by the R Consortium R Submissions Working Group for the Pilot 2.
-What’s next?
-Pilot 3
-This pilot was successfully submitted to FDA on Aug 28, 2023. This was the first publicly available R submission that included R scripts to produce ADaM datasets and TLFs. Both the ADaMs (SDTM .xpt sources from the CDISC Pilot study) and the TLFs (ADaMs .xpt sourced from the ADaMs generated in R by the Pilot 3 team) were created using R. The next steps for this pilot is to await FDA’s review and approval which may take several months to complete.
-Pilot 4
-This pilot aims to explore using technologies such as containers and WebAssembly software to package a Shiny application into a self-contained unit, streamlining the transfer and execution process for enhanced efficiency.
-This pilot is expected to be divided into two parallel submissions:
-(a) will investigate WebAssembly and
-(b) will investigate containers.
-The Journey with WebAssembly and Containers
-Our team at Appsilon teamed up with the dynamic Pilot 4 crew to explore WebAssembly technology and containers. George Stagg and Winston Chang also joined the working group to discuss the web-assembly portion of Pilot 4. This partnership brought together our engineering prowess to contribute to these tools, injecting fresh perspectives into the ongoing pilot project.
-Some of the outcomes of the collaboration:
--
-
We were able to set up a robust container environment for this pilot project.
-We aided the progress made on the use of both experimental technologies: containers and WebAssembly.
-We developed a working prototype submission using Podman container technology.
-We developed a working early-stage prototype for wrapping a small Shiny application using WebAssembly.
-
WebAssembly
-
WebAssembly allows languages like R to be executed at near-native speed directly within web browsers, providing users with the ability to run R code without having R installed locally. WebR is essentially the R programming language adapted to run in a web browser environment using WebAssembly. This project is under active development.
-The Pilot 4 Shiny App Up and Running on webR!
-
The deployed example of the Shiny app running on webR is available here. Check out the video of the application running below.
- -During this pilot, engineers at Appsilon developed a prototype of a Shiny application running on webR. The application reuses most of the code from the previous pilot apps with some tweaks and a couple of hacks/changes to get around non CRAN dependencies, specially for data loading, WebR compatibilities, and shimming some of the functionality from {teal} and other packages that are (for now) not available on CRAN.
-webR Shiny App
-During the second iteration, which was recently held, Pedro Silva shared the process of developing this Shiny app running on webR.
-The Process
--
-
Leverage last 2 iterations of the application
--
-
Reuse as much code as possible
-Avoid touching the logic part
-
-Restrict the number of dependencies to packages on CRAN
--
-
- Replace/shim functionality that was lost from removing dependencies -
-
Here is the list of dependencies to packages on CRAN, those that worked are colored green, those that were removed are marked in orange. We ended up with just 3 problematic dependencies (bold).
-| | |
-|------------------------------------|------------------------------------|
-| [library(config)]{style="color:orange"} | [library(reactable)]{style="color:green"} |
-| **library(cowplot)** | [library(rhino)]{style="color:orange"} |
-| [library(dplyr)]{style="color:green"} | [library(rtables)]{style="color:green"} |
-| [library(emmeans)]{style="color:green"} | [library(shiny)]{style="color:green"} |
-| [library(ggplot2)]{style="color:green"} | [library(stringr)]{style="color:green"} |
-| [library(glue)]{style="color:green"} | **library(teal)** |
-| [library(haven)]{style="color:green"} | **library(teal.data)** |
-| [library(htmltools)]{style="color:green"} | [library(tibble)]{style="color:green"} |
-| [library(huxtable)]{style="color:green"} | [library(tidyr)]{style="color:green"} |
-| [library(magrittr)]{style="color:green"} | [library(tippy)]{style="color:green"} |
-| [library(markdown)]{style="color:green"} | [library(Tplyr)]{style="color:green"} |
-| [library(pkgload)]{style="color:green"} | [library(utils)]{style="color:green"} |
-| [library(purrr)]{style="color:green"} | [library(visR)]{style="color:green"} |Issues with library(cowplot):
--
-
- Some issue with low-level dependencies when deployed -
Solution:
--
-
- Replace functionality with HTML -
Issues with library(teal):
--
-
- Uses {shiny.widgets} (not working for webR) -
Solution:
--
-
Redo the UI
-Load modules directly
-Recreate filter functionality
-
Issues with library(teal.data):
--
-
- Use rds exports -
Solution:
--
-
- Shim functionality, load data directly -
-
-
Leverage shinylive and httpuv to export and serve the application
--
-
Shinylive can help streamline the export process
--
-
Problems
--
-
shiny.live won’t let us have non-R files in the application directory - this is an outstanding bug that George asked us to raise an issue for.
-We wouldn’t be able to run the application as a traditional shiny app
-
-Solution:
-
1. **Custom build script**
-{httpuv} can help serve the application
--
-
- {httpuv} would run natively on a machine to serve the Shiny app -
-
-
Application Structure
-The figure below shows an overview of what we ended with:
-
Some of the issues and solutions found at the very beginning:
--
-
- The previous applications were built using golem and another one in Rhino; the support for these frameworks is not great in webR up to now.
-
-
-
- Solution
-
-
-
- {box} works out of the box (reuse the rhino version modules) -
- Simplify the structure and use a simple shiny modular structure -
-
- - Solution
-
- Shinylive does not like non-R files when generating the bundle
-
-
-
- Solution
-
-
-
- Keep the app folder as clean as possible for now (www folder only) -
-
- - Solution
-
- {teal} and {teal.data} are not on CRAN
-
-
-
- Solution
-
-
-
- Shim and used functionality -
- Use a simple tab system for the UI structure -
-
- - Solution
-
The FDA were previously told that the shiny application being prepared for the Pilot 4 submission would not be a 1 to 1 mapping from the previous one submitted for the Pilot 2 due to certain constraints such as {teal} not being on CRAN; however, this didn’t represent a problem for them since they would mainly like to test the technology.
-Pedro Silva, one of the engineers working on the development of this app mentioned “While WebR is still in development, it shows tremendous promise! The loading is definitely still a pain point (over 100mb to set up the environment!) but it will only get better moving forward.”
-Containers
-
Containerization, particularly through technologies like Docker, Podman or Singularity, offers several advantages for deploying Shiny apps.
-Choosing the Right Container
-Choosing the right container was a question that arose in this project. Although Docker is the most popular, we decided to move forward with Podman.
-In our exploration of containerization tools for deploying Shiny applications, we’ve identified key distinctions between Docker and Podman that influenced our choice.
-Podman stands out for its daemonless architecture, enhancing security by eliminating the need for a central daemon process. Unlike Docker, Podman supports running containers as non-root users, a critical feature for meeting FDA reviewer requirements. Developed by Red Hat and maintained as an open-source project, Podman prioritizes security with its rootless container support, offering a robust solution for security-conscious users.
-Goals
-A Container-based method to deploy Pilot 2 Shiny App.
-What we did
--
-
Configurable Podman Dockerfile / docker-compose.yml
--
-
- R version -
- Registry / organization name / image name (differences between docker.io and ghcr.io) -
-Documentation on creating the container
-CI: Automated build on amd64 and arm64 platforms
-
Podman short-demo
- -Below is the dockerfile (recipe) for the container:
-
Next Steps
-The next steps are waiting for the review of the Pilot 3 by the FDA and to submit the two sections that explore the new technologies to regulatory authorities. Thanks to the collaboration between the R Submission Working Group and other institutions, there is already a working prototype of a {teal}-like Shiny application running on webR and further exploration with Podman is underway.
--
-
- Submission to FDA -
- Rhino Compatibility -

Appsilon is working on the side with Rhino compatibility, eventually we might be able to just add this framework into the Pilot 4 application.
--
-
{teal}
-This and other packages might be on CRAN soon. We could incorporate them after that, replacing the shims created for this version
-Boot Time
-We need to improve boot time (remove dependencies and keep working on webR).
-
Reuse
Citation
@online{veríssimo2023,
- author = {Veríssimo, André and Makowski, Tymoteusz and Silva, Pedro
- and Viyash, Vedha and , APPSILON},
- title = {Testing {Containers} and {WebAssembly} in {Submissions} to
- the {FDA}},
- date = {2023-02-01},
- url = {https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html},
- langid = {en}
-}
-Conclusion
Last updated
2024-02-01 12:24:22.445948
+2024-02-01 12:42:29.277076
Details
Compare
Last updated
2024-02-01 12:24:16.723448
+2024-02-01 12:42:23.618603
Details
Conclusion
Last updated
2024-02-01 12:24:14.841985
+2024-02-01 12:42:21.757191
Details
How can I make
Last updated
-2024-02-01 12:24:31.481975
+2024-02-01 12:42:38.30988
Details
diff --git a/posts/2023-07-09_falcon/falcon.html b/posts/2023-07-09_falcon/falcon.html
index f9f74824..31ad46f4 100644
--- a/posts/2023-07-09_falcon/falcon.html
+++ b/posts/2023-07-09_falcon/falcon.html
@@ -207,7 +207,7 @@ Next steps & vision<
Last updated
-2024-02-01 12:24:34.960458
+2024-02-01 12:42:41.841083
Details
diff --git a/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html b/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
index 3a876286..b96da499 100644
--- a/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
+++ b/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
@@ -312,7 +312,7 @@ That’s it!
Last updated
-2024-02-01 12:24:09.63242
+2024-02-01 12:42:16.798817
Details
diff --git a/posts/2023-07-14_code_sections/code_sections.html b/posts/2023-07-14_code_sections/code_sections.html
index ef0829a3..d3f84894 100644
--- a/posts/2023-07-14_code_sections/code_sections.html
+++ b/posts/2023-07-14_code_sections/code_sections.html
@@ -305,7 +305,7 @@ Conclusion
Last updated
-2024-02-01 12:24:18.44166
+2024-02-01 12:42:25.336389
Details
diff --git a/posts/2023-07-24_rounding/rounding.html b/posts/2023-07-24_rounding/rounding.html
index 90e64b41..b1952e33 100644
--- a/posts/2023-07-24_rounding/rounding.html
+++ b/posts/2023-07-24_rounding/rounding.html
@@ -478,7 +478,7 @@ Conclusion
Last updated
-2024-02-01 12:24:20.344918
+2024-02-01 12:42:27.242831
Details
diff --git a/posts/2023-08-08_study_day/study_day.html b/posts/2023-08-08_study_day/study_day.html
index 700db0d9..60db0b48 100644
--- a/posts/2023-08-08_study_day/study_day.html
+++ b/posts/2023-08-08_study_day/study_day.html
@@ -275,7 +275,7 @@ It’s all relative? - Calculating Relative Days using admiral
Last updated
-2024-02-01 12:24:33.442683
+2024-02-01 12:42:40.324505
Details
diff --git a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
index 1a3fdf99..3bf33dbd 100644
--- a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+++ b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
@@ -376,7 +376,7 @@ References
Last updated
-2024-02-01 12:24:41.550192
+2024-02-01 12:42:47.195254
Details
diff --git a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
index 08b1a0a8..335ca2a4 100644
--- a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+++ b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
@@ -232,7 +232,7 @@ Release, g
Last updated
-2024-02-01 12:24:26.757735
+2024-02-01 12:42:33.536958
Details
diff --git a/posts/2023-10-30_floating_point/floating_point.html b/posts/2023-10-30_floating_point/floating_point.html
index e0c45bd8..ba3d6404 100644
--- a/posts/2023-10-30_floating_point/floating_point.html
+++ b/posts/2023-10-30_floating_point/floating_point.html
@@ -340,8 +340,8 @@ Issues arising
select(TERM, GRADE_CRITERIA_CODE) %>%
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
@@ -598,8 +598,8 @@ Potential solutions select(TERM, GRADE_CRITERIA_CODE) %>%
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
Conclusion
Last updated
-2024-02-01 12:24:43.976417
+2024-02-01 12:42:49.540771
Details
diff --git a/posts/2023-11-27_higher_order/higher_order.html b/posts/2023-11-27_higher_order/higher_order.html
index d3425d57..153e4a1f 100644
--- a/posts/2023-11-27_higher_order/higher_order.html
+++ b/posts/2023-11-27_higher_order/higher_order.html
@@ -804,7 +804,7 @@ Conclusion
Last updated
-2024-02-01 12:24:06.874564
+2024-02-01 12:42:12.894734
Details
diff --git a/posts/2023-12-18_admiral_1_0/admiral_1_0.html b/posts/2023-12-18_admiral_1_0/admiral_1_0.html
index 63fc3750..858d190f 100644
--- a/posts/2023-12-18_admiral_1_0/admiral_1_0.html
+++ b/posts/2023-12-18_admiral_1_0/admiral_1_0.html
@@ -509,7 +509,7 @@ Way Back Machine
Last updated
-2024-02-01 12:24:37.177593
+2024-02-01 12:42:44.050491
Details
diff --git a/posts/2023-12-20_p_k__examples/p_k__examples.html b/posts/2023-12-20_p_k__examples/p_k__examples.html
index 00b7e9ef..de011ee7 100644
--- a/posts/2023-12-20_p_k__examples/p_k__examples.html
+++ b/posts/2023-12-20_p_k__examples/p_k__examples.html
@@ -373,7 +373,7 @@ Derive Additio
Last updated
-2024-02-01 12:24:29.879149
+2024-02-01 12:42:36.670717
Details
diff --git a/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html b/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
index 901be65d..700d19d5 100644
--- a/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
+++ b/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
@@ -208,7 +208,7 @@ P.S.
Last updated
-2024-02-01 12:24:38.870695
+2024-02-01 12:42:45.713213
Details
diff --git a/posts/2023-12-30_containers_webassembly_submission/FDA_WebR_Rhino.webp b/posts/2024-02-01_containers_webassembly_submission/FDA_WebR_Rhino.webp
similarity index 100%
rename from posts/2023-12-30_containers_webassembly_submission/FDA_WebR_Rhino.webp
rename to posts/2024-02-01_containers_webassembly_submission/FDA_WebR_Rhino.webp
diff --git a/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html b/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
similarity index 98%
rename from posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html
rename to posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
index feda2cd7..d157ad8d 100644
--- a/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html
+++ b/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
@@ -11,7 +11,7 @@
-
+
Pharmaverse Blog - Testing Containers and WebAssembly in Submissions to the FDA
@@ -86,7 +86,7 @@
-
+
@@ -154,7 +154,7 @@ Testing Containers and WebAssembly in Submissions to the FDA
@@ -436,19 +436,19 @@ Next Steps
Reuse
Citation
BibTeX citation:@online{veríssimo2023,
+Reuse
Citation
BibTeX citation:@online{veríssimo2024,
author = {Veríssimo, André and Makowski, Tymoteusz and Silva, Pedro
and Viyash, Vedha and , APPSILON},
title = {Testing {Containers} and {WebAssembly} in {Submissions} to
the {FDA}},
- date = {2023-02-01},
- url = {https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html},
+ date = {2024-02-01},
+ url = {https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html},
langid = {en}
}
-
For attribution, please cite this work as:
+For attribution, please cite this work as:
Veríssimo, André, Tymoteusz Makowski, Pedro Silva, Vedha Viyash, and
-APPSILON. 2023. “Testing Containers and WebAssembly in Submissions
-to the FDA.” February 1, 2023. https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html.
+APPSILON. 2024. “Testing Containers and WebAssembly in Submissions
+to the FDA.” February 1, 2024. https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html.
Last updated
2024-02-01 12:24:31.481975
+2024-02-01 12:42:38.30988
Details
Next steps & vision<
Last updated
-2024-02-01 12:24:34.960458
+2024-02-01 12:42:41.841083
Details
diff --git a/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html b/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
index 3a876286..b96da499 100644
--- a/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
+++ b/posts/2023-07-10_blanks_and_nas/blanks_and_nas.html
@@ -312,7 +312,7 @@ That’s it!
Last updated
-2024-02-01 12:24:09.63242
+2024-02-01 12:42:16.798817
Details
diff --git a/posts/2023-07-14_code_sections/code_sections.html b/posts/2023-07-14_code_sections/code_sections.html
index ef0829a3..d3f84894 100644
--- a/posts/2023-07-14_code_sections/code_sections.html
+++ b/posts/2023-07-14_code_sections/code_sections.html
@@ -305,7 +305,7 @@ Conclusion
Last updated
-2024-02-01 12:24:18.44166
+2024-02-01 12:42:25.336389
Details
diff --git a/posts/2023-07-24_rounding/rounding.html b/posts/2023-07-24_rounding/rounding.html
index 90e64b41..b1952e33 100644
--- a/posts/2023-07-24_rounding/rounding.html
+++ b/posts/2023-07-24_rounding/rounding.html
@@ -478,7 +478,7 @@ Conclusion
Last updated
-2024-02-01 12:24:20.344918
+2024-02-01 12:42:27.242831
Details
diff --git a/posts/2023-08-08_study_day/study_day.html b/posts/2023-08-08_study_day/study_day.html
index 700db0d9..60db0b48 100644
--- a/posts/2023-08-08_study_day/study_day.html
+++ b/posts/2023-08-08_study_day/study_day.html
@@ -275,7 +275,7 @@ It’s all relative? - Calculating Relative Days using admiral
Last updated
-2024-02-01 12:24:33.442683
+2024-02-01 12:42:40.324505
Details
diff --git a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
index 1a3fdf99..3bf33dbd 100644
--- a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+++ b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
@@ -376,7 +376,7 @@ References
Last updated
-2024-02-01 12:24:41.550192
+2024-02-01 12:42:47.195254
Details
diff --git a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
index 08b1a0a8..335ca2a4 100644
--- a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+++ b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
@@ -232,7 +232,7 @@ Release, g
Last updated
-2024-02-01 12:24:26.757735
+2024-02-01 12:42:33.536958
Details
diff --git a/posts/2023-10-30_floating_point/floating_point.html b/posts/2023-10-30_floating_point/floating_point.html
index e0c45bd8..ba3d6404 100644
--- a/posts/2023-10-30_floating_point/floating_point.html
+++ b/posts/2023-10-30_floating_point/floating_point.html
@@ -340,8 +340,8 @@ Issues arising
select(TERM, GRADE_CRITERIA_CODE) %>%
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
@@ -598,8 +598,8 @@ Potential solutions select(TERM, GRADE_CRITERIA_CODE) %>%
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
Conclusion
Last updated
-2024-02-01 12:24:43.976417
+2024-02-01 12:42:49.540771
Details
diff --git a/posts/2023-11-27_higher_order/higher_order.html b/posts/2023-11-27_higher_order/higher_order.html
index d3425d57..153e4a1f 100644
--- a/posts/2023-11-27_higher_order/higher_order.html
+++ b/posts/2023-11-27_higher_order/higher_order.html
@@ -804,7 +804,7 @@ Conclusion
Last updated
-2024-02-01 12:24:06.874564
+2024-02-01 12:42:12.894734
Details
diff --git a/posts/2023-12-18_admiral_1_0/admiral_1_0.html b/posts/2023-12-18_admiral_1_0/admiral_1_0.html
index 63fc3750..858d190f 100644
--- a/posts/2023-12-18_admiral_1_0/admiral_1_0.html
+++ b/posts/2023-12-18_admiral_1_0/admiral_1_0.html
@@ -509,7 +509,7 @@ Way Back Machine
Last updated
-2024-02-01 12:24:37.177593
+2024-02-01 12:42:44.050491
Details
diff --git a/posts/2023-12-20_p_k__examples/p_k__examples.html b/posts/2023-12-20_p_k__examples/p_k__examples.html
index 00b7e9ef..de011ee7 100644
--- a/posts/2023-12-20_p_k__examples/p_k__examples.html
+++ b/posts/2023-12-20_p_k__examples/p_k__examples.html
@@ -373,7 +373,7 @@ Derive Additio
Last updated
-2024-02-01 12:24:29.879149
+2024-02-01 12:42:36.670717
Details
diff --git a/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html b/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
index 901be65d..700d19d5 100644
--- a/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
+++ b/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
@@ -208,7 +208,7 @@ P.S.
Last updated
-2024-02-01 12:24:38.870695
+2024-02-01 12:42:45.713213
Details
diff --git a/posts/2023-12-30_containers_webassembly_submission/FDA_WebR_Rhino.webp b/posts/2024-02-01_containers_webassembly_submission/FDA_WebR_Rhino.webp
similarity index 100%
rename from posts/2023-12-30_containers_webassembly_submission/FDA_WebR_Rhino.webp
rename to posts/2024-02-01_containers_webassembly_submission/FDA_WebR_Rhino.webp
diff --git a/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html b/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
similarity index 98%
rename from posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html
rename to posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
index feda2cd7..d157ad8d 100644
--- a/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html
+++ b/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
@@ -11,7 +11,7 @@
-
+
Pharmaverse Blog - Testing Containers and WebAssembly in Submissions to the FDA
@@ -86,7 +86,7 @@
-
+
@@ -154,7 +154,7 @@ Testing Containers and WebAssembly in Submissions to the FDA
@@ -436,19 +436,19 @@ Next Steps
Reuse
Citation
BibTeX citation:@online{veríssimo2023,
+Reuse
Citation
BibTeX citation:@online{veríssimo2024,
author = {Veríssimo, André and Makowski, Tymoteusz and Silva, Pedro
and Viyash, Vedha and , APPSILON},
title = {Testing {Containers} and {WebAssembly} in {Submissions} to
the {FDA}},
- date = {2023-02-01},
- url = {https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html},
+ date = {2024-02-01},
+ url = {https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html},
langid = {en}
}
-
For attribution, please cite this work as:
+For attribution, please cite this work as:
Veríssimo, André, Tymoteusz Makowski, Pedro Silva, Vedha Viyash, and
-APPSILON. 2023. “Testing Containers and WebAssembly in Submissions
-to the FDA.” February 1, 2023. https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html.
+APPSILON. 2024. “Testing Containers and WebAssembly in Submissions
+to the FDA.” February 1, 2024. https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html.
Last updated
2024-02-01 12:24:34.960458
+2024-02-01 12:42:41.841083
Details
That’s it!
Last updated
2024-02-01 12:24:09.63242
+2024-02-01 12:42:16.798817
Details
Conclusion
Last updated
2024-02-01 12:24:18.44166
+2024-02-01 12:42:25.336389
Details
Conclusion
Last updated
2024-02-01 12:24:20.344918
+2024-02-01 12:42:27.242831
Details
It’s all relative? - Calculating Relative Days using admiral
Last updated
-2024-02-01 12:24:33.442683
+2024-02-01 12:42:40.324505
Details
diff --git a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
index 1a3fdf99..3bf33dbd 100644
--- a/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
+++ b/posts/2023-08-14_rhino_submission_2/rhino_submission_2.html
@@ -376,7 +376,7 @@ References
Last updated
-2024-02-01 12:24:41.550192
+2024-02-01 12:42:47.195254
Details
diff --git a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
index 08b1a0a8..335ca2a4 100644
--- a/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
+++ b/posts/2023-10-10_pharmaverse_story/pharmaverse_story.html
@@ -232,7 +232,7 @@ Release, g
Last updated
-2024-02-01 12:24:26.757735
+2024-02-01 12:42:33.536958
Details
diff --git a/posts/2023-10-30_floating_point/floating_point.html b/posts/2023-10-30_floating_point/floating_point.html
index e0c45bd8..ba3d6404 100644
--- a/posts/2023-10-30_floating_point/floating_point.html
+++ b/posts/2023-10-30_floating_point/floating_point.html
@@ -340,8 +340,8 @@ Issues arising
select(TERM, GRADE_CRITERIA_CODE) %>%
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
@@ -598,8 +598,8 @@ Potential solutions select(TERM, GRADE_CRITERIA_CODE) %>%
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
Conclusion
Last updated
-2024-02-01 12:24:43.976417
+2024-02-01 12:42:49.540771
Details
diff --git a/posts/2023-11-27_higher_order/higher_order.html b/posts/2023-11-27_higher_order/higher_order.html
index d3425d57..153e4a1f 100644
--- a/posts/2023-11-27_higher_order/higher_order.html
+++ b/posts/2023-11-27_higher_order/higher_order.html
@@ -804,7 +804,7 @@ Conclusion
Last updated
-2024-02-01 12:24:06.874564
+2024-02-01 12:42:12.894734
Details
diff --git a/posts/2023-12-18_admiral_1_0/admiral_1_0.html b/posts/2023-12-18_admiral_1_0/admiral_1_0.html
index 63fc3750..858d190f 100644
--- a/posts/2023-12-18_admiral_1_0/admiral_1_0.html
+++ b/posts/2023-12-18_admiral_1_0/admiral_1_0.html
@@ -509,7 +509,7 @@ Way Back Machine
Last updated
-2024-02-01 12:24:37.177593
+2024-02-01 12:42:44.050491
Details
diff --git a/posts/2023-12-20_p_k__examples/p_k__examples.html b/posts/2023-12-20_p_k__examples/p_k__examples.html
index 00b7e9ef..de011ee7 100644
--- a/posts/2023-12-20_p_k__examples/p_k__examples.html
+++ b/posts/2023-12-20_p_k__examples/p_k__examples.html
@@ -373,7 +373,7 @@ Derive Additio
Last updated
-2024-02-01 12:24:29.879149
+2024-02-01 12:42:36.670717
Details
diff --git a/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html b/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
index 901be65d..700d19d5 100644
--- a/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
+++ b/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
@@ -208,7 +208,7 @@ P.S.
Last updated
-2024-02-01 12:24:38.870695
+2024-02-01 12:42:45.713213
Details
diff --git a/posts/2023-12-30_containers_webassembly_submission/FDA_WebR_Rhino.webp b/posts/2024-02-01_containers_webassembly_submission/FDA_WebR_Rhino.webp
similarity index 100%
rename from posts/2023-12-30_containers_webassembly_submission/FDA_WebR_Rhino.webp
rename to posts/2024-02-01_containers_webassembly_submission/FDA_WebR_Rhino.webp
diff --git a/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html b/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
similarity index 98%
rename from posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html
rename to posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
index feda2cd7..d157ad8d 100644
--- a/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html
+++ b/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
@@ -11,7 +11,7 @@
-
+
Pharmaverse Blog - Testing Containers and WebAssembly in Submissions to the FDA
@@ -86,7 +86,7 @@
-
+
@@ -154,7 +154,7 @@ Testing Containers and WebAssembly in Submissions to the FDA
@@ -436,19 +436,19 @@ Next Steps
Reuse
Citation
BibTeX citation:@online{veríssimo2023,
+Reuse
Citation
BibTeX citation:@online{veríssimo2024,
author = {Veríssimo, André and Makowski, Tymoteusz and Silva, Pedro
and Viyash, Vedha and , APPSILON},
title = {Testing {Containers} and {WebAssembly} in {Submissions} to
the {FDA}},
- date = {2023-02-01},
- url = {https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html},
+ date = {2024-02-01},
+ url = {https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html},
langid = {en}
}
-
For attribution, please cite this work as:
+For attribution, please cite this work as:
Veríssimo, André, Tymoteusz Makowski, Pedro Silva, Vedha Viyash, and
-APPSILON. 2023. “Testing Containers and WebAssembly in Submissions
-to the FDA.” February 1, 2023. https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html.
+APPSILON. 2024. “Testing Containers and WebAssembly in Submissions
+to the FDA.” February 1, 2024. https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html.
Last updated
2024-02-01 12:24:33.442683
+2024-02-01 12:42:40.324505
Details
References
Last updated
2024-02-01 12:24:41.550192
+2024-02-01 12:42:47.195254
Details
Release, g
Last updated
-2024-02-01 12:24:26.757735
+2024-02-01 12:42:33.536958
Details
diff --git a/posts/2023-10-30_floating_point/floating_point.html b/posts/2023-10-30_floating_point/floating_point.html
index e0c45bd8..ba3d6404 100644
--- a/posts/2023-10-30_floating_point/floating_point.html
+++ b/posts/2023-10-30_floating_point/floating_point.html
@@ -340,8 +340,8 @@ Issues arising
select(TERM, GRADE_CRITERIA_CODE) %>%
reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
-
-
+
+
Last updated
2024-02-01 12:24:26.757735
+2024-02-01 12:42:33.536958
Details
Issues arising
select(TERM, GRADE_CRITERIA_CODE) %>% reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)As you can see, the data-frame contains the column GRADE_CRITERIA_CODE which contains comparisons of floating point values. And there was a discrepancy of what Gordon expected to see, and how R actually computed the comparison initially:
Potential solutions select(TERM, GRADE_CRITERIA_CODE) %>% reactable(defaultPageSize = 4, highlight = TRUE, bordered = TRUE, striped = TRUE, resizable = TRUE)
Conclusion
Last updated
2024-02-01 12:24:43.976417
+2024-02-01 12:42:49.540771
Details
Conclusion
Last updated
2024-02-01 12:24:06.874564
+2024-02-01 12:42:12.894734
Details
Way Back Machine
Last updated
2024-02-01 12:24:37.177593
+2024-02-01 12:42:44.050491
Details
Derive Additio
Last updated
-2024-02-01 12:24:29.879149
+2024-02-01 12:42:36.670717
Details
diff --git a/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html b/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
index 901be65d..700d19d5 100644
--- a/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
+++ b/posts/2024-01-04_end_of__year__up.../end_of__year__update_from_the__pharmaverse__council.html
@@ -208,7 +208,7 @@ P.S.
Last updated
-2024-02-01 12:24:38.870695
+2024-02-01 12:42:45.713213
Details
diff --git a/posts/2023-12-30_containers_webassembly_submission/FDA_WebR_Rhino.webp b/posts/2024-02-01_containers_webassembly_submission/FDA_WebR_Rhino.webp
similarity index 100%
rename from posts/2023-12-30_containers_webassembly_submission/FDA_WebR_Rhino.webp
rename to posts/2024-02-01_containers_webassembly_submission/FDA_WebR_Rhino.webp
diff --git a/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html b/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
similarity index 98%
rename from posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html
rename to posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
index feda2cd7..d157ad8d 100644
--- a/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html
+++ b/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html
@@ -11,7 +11,7 @@
-
+
Pharmaverse Blog - Testing Containers and WebAssembly in Submissions to the FDA
@@ -86,7 +86,7 @@
-
+
@@ -154,7 +154,7 @@ Testing Containers and WebAssembly in Submissions to the FDA
@@ -436,19 +436,19 @@ Next Steps
Reuse
Citation
BibTeX citation:@online{veríssimo2023,
+Reuse
Citation
BibTeX citation:@online{veríssimo2024,
author = {Veríssimo, André and Makowski, Tymoteusz and Silva, Pedro
and Viyash, Vedha and , APPSILON},
title = {Testing {Containers} and {WebAssembly} in {Submissions} to
the {FDA}},
- date = {2023-02-01},
- url = {https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html},
+ date = {2024-02-01},
+ url = {https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html},
langid = {en}
}
-
For attribution, please cite this work as:
+For attribution, please cite this work as:
Veríssimo, André, Tymoteusz Makowski, Pedro Silva, Vedha Viyash, and
-APPSILON. 2023. “Testing Containers and WebAssembly in Submissions
-to the FDA.” February 1, 2023. https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html.
+APPSILON. 2024. “Testing Containers and WebAssembly in Submissions
+to the FDA.” February 1, 2024. https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html.
Last updated
2024-02-01 12:24:29.879149
+2024-02-01 12:42:36.670717
Details
P.S.
Last updated
2024-02-01 12:24:38.870695
+2024-02-01 12:42:45.713213
Details
Testing Containers and WebAssembly in Submissions to the FDA
Next Steps
Reuse
Citation
@online{veríssimo2023,
+Reuse
Citation
BibTeX citation:@online{veríssimo2024,
author = {Veríssimo, André and Makowski, Tymoteusz and Silva, Pedro
and Viyash, Vedha and , APPSILON},
title = {Testing {Containers} and {WebAssembly} in {Submissions} to
the {FDA}},
- date = {2023-02-01},
- url = {https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html},
+ date = {2024-02-01},
+ url = {https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html},
langid = {en}
}
-
For attribution, please cite this work as:
+For attribution, please cite this work as:
Veríssimo, André, Tymoteusz Makowski, Pedro Silva, Vedha Viyash, and
-APPSILON. 2023. “Testing Containers and WebAssembly in Submissions
-to the FDA.” February 1, 2023. https://pharmaverse.github.io/blog/posts/2023-12-30_containers_webassembly_submission/containers_and_webassembly_submissions.html.
+APPSILON. 2024. “Testing Containers and WebAssembly in Submissions
+to the FDA.” February 1, 2024. https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html.

















