-
Notifications
You must be signed in to change notification settings - Fork 12
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Casey Kneale
committed
Oct 17, 2020
1 parent
d8cd288
commit 397b91b
Showing
5 changed files
with
68 additions
and
12 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,3 @@ | ||
| { | ||
| "julia.environmentPath": "/home/caseykneale/Desktop/ChemometricsTools.jl" | ||
| } |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,49 @@ | ||
| # Other Packages: | ||
| So you know what you're doing, you're not one of those Friday night chemometricians Brereton talked about, and you want to compare some methods available in ChemometricsTools.jl. Great! The nice thing about Julia is, packages tend to work with one another with zero effort. To demonstrate this I made a little tutorial using (Turing.jl)[https://turing.ml/dev/] and (ChemometricsData.jl)[https://github.com/caseykneale/ChemometricsData.jl] for a very basic incomplete analysis of some well known bayesian regression methods. Let's get started. | ||
|
|
||
|
|
||
| ## Lets load in some data | ||
| ```julia | ||
| using Turing, StatsPlots, Plots, Statistics | ||
| using DataFrames, ChemometricsData | ||
|
|
||
| println( ChemometricsData.search("corn") ) | ||
| corn_data = ChemometricsData.load("Cargill_Corn") | ||
| X = Matrix(corn_data["m5_spectra.csv"]) | ||
|
|
||
| xaxis = 1100:2:2498#nm | ||
|
|
||
| plot( X', title = "Cargill Corn M5 Spec", xlab = "Wavelength (nm)", ylab = "Absorbance", legend = false, | ||
| xticks = (1:50:length(xaxis), xaxis[1:50:end]) ) | ||
| ``` | ||
|  | ||
|
|
||
| Grab our property values, | ||
| ```julia | ||
| Y = corn_data["property_values.csv"][!,:Moisture] | ||
| ``` | ||
|
|
||
| Now let's center and scale our X and Y values to keep our regression methods happy | ||
|
|
||
| ```julia | ||
| train, test = 1:35, 36:80 | ||
| X_train, X_test = X_processed[train,:], X_processed[test,:] | ||
| μx,σx = mean(X_train, dims = 1), std(X_train, dims = 1) | ||
| X_train = (X_train .- μx) ./ σx | ||
| X_test = (X_test .- μx) ./ σx | ||
|
|
||
| Y_train, Y_test = Y[train,:], Y[test,:] | ||
| μy,σy = mean(Y_train),std(Y_train) | ||
| Y_train = (Y_train .- μy) ./ σy | ||
| Y_test = (Y_test .- μy) ./ σy | ||
| ``` | ||
|
|
||
|
|
||
|  | ||
|
|
||
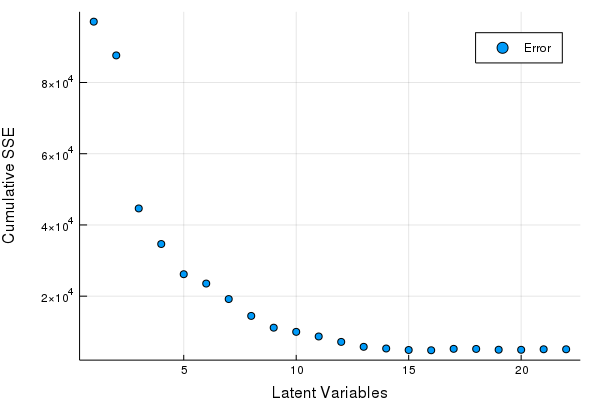
| That's great right? but, hey that was kind of slow. Knowing what we know about ALS based models, we can do the same operation in linear time with respect to latent factors by computing the most latent variables first and only recomputing the regression coefficients. An example of this is below, | ||
|
|
||
| ```julia | ||
|
|
||
| ``` | ||
| This approach is ~5 times faster on a single core( < 2 seconds), pours through 7Gb less data, and makes 1/5th the allocations (on this dataset at least). If you wanted you could distribute the inner loop (using Distributed.jl) and see drastic speed ups! |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters