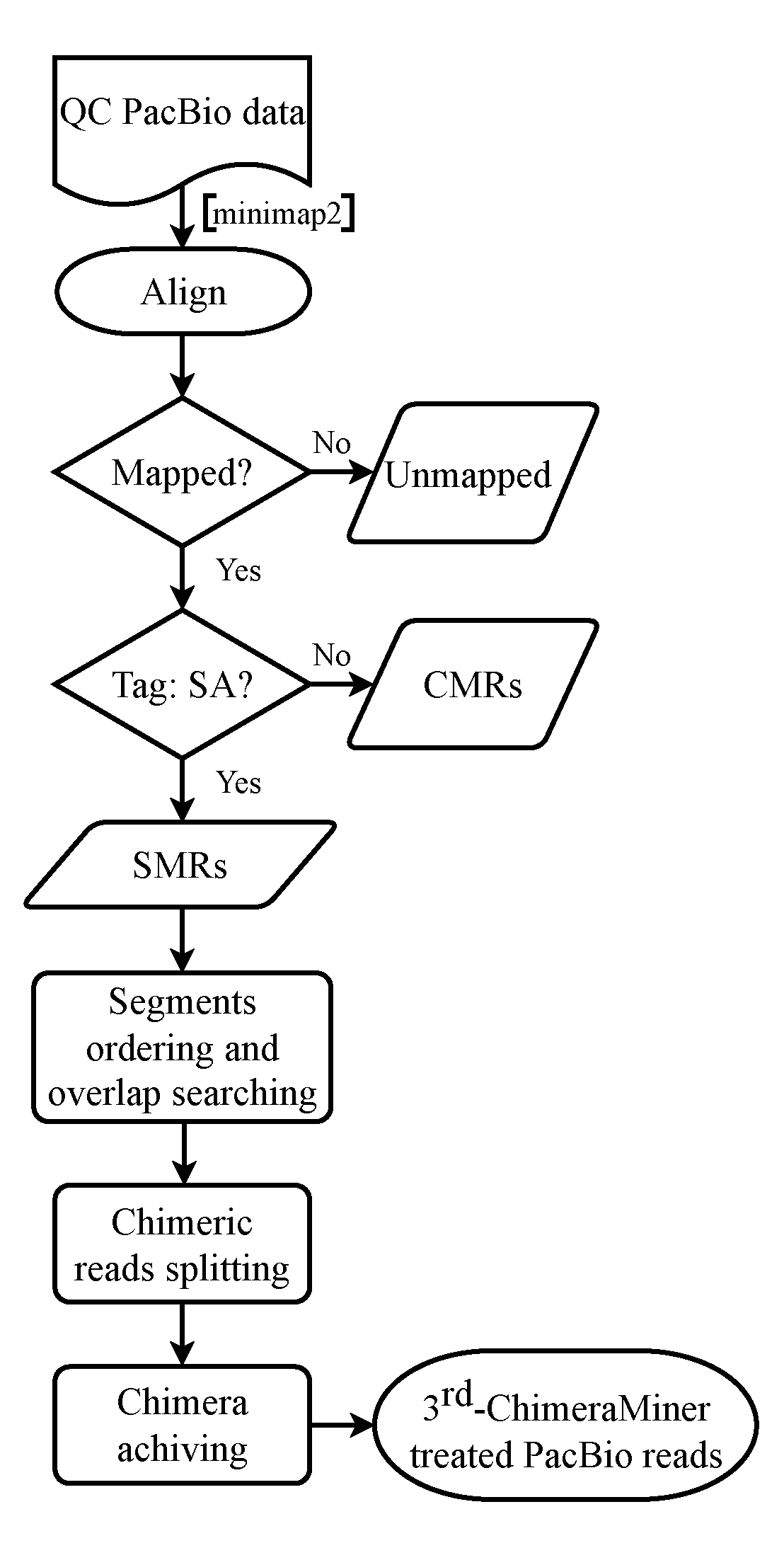
Recognizing chimeras in long-read sequencing data and converting chimeric reads into normal and full-length mappable reads.
Exploration of whole genome amplification generated chimeric sequences in long-read sequencing data.
The pipeline of the 3rd-ChimeraMiner is following as:
- perl (version: v5.22.1) built for x86_64-linux-gnu-thread-multi
- minimap2 (version: 2.22-r1105-dirty)
- samtools (version: 1.11-5-g0920974), Using htslib 1.11-18-g7f8211c
- sambamba (version: 0.8.0), Using LDC 1.24.0 / DMD v2.094.1 / LLVM11.0.0 / bootstrap LDC - the LLVM D compiler (1.24.0)
- SMRT Link (version: 10.2.0.133434)
Install these softwares and add the PATH to the environment PATH, for example:
echo export PATH=/the/fold/of/the/executable/file:\$PATH >> ~/.bashrc
source ~/.bashrc
# In my environment, the path of these softwares
grep -E 'samtools|sambamba|minimap2|htslib|smart' ~/.bashrc
PATH=/home/luna/Desktop/Software/sambamba/build:$PATH
PATH=/home/luna/Desktop/Software/samtools/samtools:$PATH
PATH=/home/luna/Desktop/Software/minimap2:$PATH
PATH=/home/luna/Desktop/Software/samtools/htslib/bin:$PATH
PATH=/home/luna/Desktop/Software/PacificBiosciences/smrtlink/admin/bin:$PATH
PATH=/home/luna/Desktop/Software/PacificBiosciences/smrtlink/smrtcmds/developer/bin:$PATH
PATH=/home/luna/Desktop/Software/PacificBiosciences/smrtlink/smrtcmds/bin:$PATHIn this pipeline, we need some Perl Modules, we should install these modules before run it.
- Getopt::Long
- Cwd qw(abs_path)
- File::Basename
- File::Spec
I recommand to use "cpanm" to install Perl Modules.
curl -L https://cpanmin.us | perl - App::cpanminus
echo "eval \`perl -I ~/perl5/lib/perl5 -Mlocal::lib\`" >> ~/.bashrc
source ~/.bashrc
cpanm -v --notest -l ~/perl5 Getopt::Long
cpanm -v --notest -l ~/perl5 File::Spec
cpanm -v --notest -l ~/perl5 File::BasenameIn my local server, the fold of the human reference genome (hg19) is:
/home/luna/Desktop/database/homo_minimap2
So, in your analysis environment , please change to your reference directory.
In this fold, we have the reference genome fasta file hsa.fa, including all sequence of all chromosome (chr{1..22, X, Y}) which downloaded from UCSC hg19, and fasta file have indexed with samtools and minimap2.
# download minimap2
curl -L https://github.com/lh3/minimap2/releases/download/v2.24/minimap2-2.24_x64-linux.tar.bz2 | tar -jxvf -
./minimap2-2.24_x64-linux/minimap2
echo "export PATH=$PWD/minimap2-2.24_x64-linux:\$PATH" >> ~/.bashrc
source ~/.bashrc
# download referenge genome sequences from UCSC
curl -L https://hgdownload.cse.ucsc.edu/goldenPath/hg19/bigZips/chromFa.tar.gz | tar -xzvf -
# delete useless
rm -rf chrUn*.fa chr*random.fa chr*hap*.fa
# index for each chromosome fasta
for i in 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 X Y M
do
samtools faidx chr${i}.fa
done
# merge chromosome fasta into a fasta as the hg19 fa, and index for reference genome sequence
cat chr1.fa chr2.fa chr3.fa chr4.fa chr5.fa chr6.fa chr7.fa chr8.fa chr9.fa chr10.fa chr11.fa chr12.fa chr13.fa chr14.fa chr15.fa chr16.fa chr17.fa chr18.fa chr19.fa chr20.fa chr21.fa chr22.fa chrX.fa chrY.fa chrM.fa > hsa.fa
samtools faidx hsa.fa
minimap2 -x map-pb -d hsa.pb.mmi hsa.fa
minimap2 -x map-hifi -d hsa.HIFI.mmi hsa.faIn here, please make sure the name of your chromosomes is like "chr1 chr2 chr3 ... chrX chrY", Thanks.
In addition, minimap2 can align DNA sequences against the reference genome sequence by using presets without indexed.
The folder contains test running shell scripts. It turns out that all the scripts are running. You can check out how to use the pipeline.
Before run the shell script, we need to download the bigger bam file and 'bam.pbi' file into the 'exampledata' folder, the download link of the example dataset is https://pan.seu.edu.cn:443/link/74E6F25A348813990FD47C480009C4D8 . Because of the dataset file is too big to upload directly to the GitHub.
If the link is expired, please do not hesitate to contact me directly by email.
In this folder, just run workstep.sh first, this shell will generate bam file and chimera's files. When all works in workstep.sh finished, then run filterstep.sh, this shell will deal with the chimera's files and count.
Use samtools fastq to convert bam file to fastq file, and use minimap2 to align the PacBio reads to hg19 and generate a aligned bam file for downstream analysis;
dir=/the/folder/of/the/3rd-ChimeraMiner
cd $dir/exampledata
ref=/home/luna/Desktop/database/homo_minimap2/hsa.fa
mmi=/home/luna/Desktop/database/homo_minimap2/hsa.HIFI.mmi
samtools=/home/luna/Desktop/Software/samtools/samtools/samtools
minimap2=/home/luna/Desktop/Software/minimap2/minimap2
samp=example
# convert the ccs bam into fastq for minimap2 as the input file
$samtools fastq -@ 20 $samp.ccs.bam | gzip > $samp.ccs.fq.gz
# align to the hg19 with indexed
minimap2 -Y -t 60 -a $mmi --cs --MD $samp.ccs.fq.gz | samtools view -Sb -@ 4 -o $samp.bam
# or align to the hg19 without indexed
minimap2 -Y -t 60 -ax map-hifi $ref --cs --MD $samp.ccs.fq.gz | samtools view -Sb -@ 4 -o $samp.bamThis step was performed by using the script ExtIDsGenShell.pl. It uses an aligned bam file as the input file. First, splitting the bam file into multiple bam files, each bam file contains "$number" reads id (default was set to 100000). Then, generate a shell script for running the 3rd-ChimeraMiner.
# Test for the small bam file in exampledata folder
perl $dir/ExtIDsGenShell.pl -i $samp.bam -d $dir/exampledata/result -m $samp -n 5000 -r $ref -sn 1
# or Test for the bigger bam file downloaded from `pan.seu.edu.cn`
perl $dir/ExtIDsGenShell.pl -i $samp.bam -d $dir/exampledata/result -m $samp -n 20000 -r $ref -sn 1
# after finished the above process
cd $dir/exampledata/result/$samp
ls
id.$samp.part.1.bam id.$samp.part.1.bam.bai id.$samp.part.2.bam id.$samp.part.2.bam.bai id.$samp.part.3.bam id.$samp.part.3.bam.bai
...After waiting for the split and generate process to finish, multiple sub-bam files will be generated and stored in $dir/exampledata/result/example and will be indexed automatically by using samtools index. In addition, a shell script example.runFind.sh will be generated and stored in $dir/exampledata/result/example
The shell script example.runFind.sh which obtained by the previous step, do four works by one Perl script: FindChimeras.latest.pl
-
Definition of continuously mapped reads (CMRs) and segmented mapped reads (SMRs) according to the alignment of read without or with “SA” tag.
-
Unscrambling of the SMRs with CIGAR and "SA" TAG information
-
Overlap searching of two adjacent segments of SMRs
-
Acquisition of the efficient chimeras
Sometimes, we recommended that split the shell script example.runFind.sh. And in this script, three lines are a process. We could split it according the feature.
cd $dir/exampledata/result/$samp
split -l 3 $samp.runFind.sh split.Find.
ls split.Find.* | grep -v log | perl -ne 'chomp;`nohup sh $_ &> $_.log &`;'This will allow you to detect chimeric reads in each sub-bam file. Just waiting for these split scripts completed and will achieve the .chimera files of chimeric reads of each sub-bam file:
ls
$samp.part.1.chimera $samp.part.2.chimera $samp.part.3.chimera ...The format of the .chimera file is following as:
First line is the information of one PacBio read:
read_id read_length read_seq
Second line is the former segment of the first pair chimeric sequence:
Seg_1 Overlap Distance chr_ref strand_ref start_ref end_ref start_in_seq end_in_seq Seg_1_sequence
Third line is the later segment of the first pair chimeric read and the former segment of the second pair chimeric sequence:
Seg_2 Overlap_information Distance chr_ref strand_ref start_ref end_ref start_in_seq end_in_seq Seg_2_sequence
.....
The rest segments were arranged according to the above rules until a new PacBio read was encountered, for example:
m64030_220717_044032/16/ccs 2210 CGAGGAAGAAGCAAGAGCAGAAACCTGGATAAACCCATCAGCTCTCATGAGACTTATTCACTATCAATGAGAATAGCATGGAAAGACCGGCCCCCATGATTCAATTACCTCCCCCTGGGTCCCTCCCACAACATATGAGAATTCTGGAGATACAATTCAAGTTGATATTTGGGTGGAGAGACAGCCAAACCATATCATACGGCCCCTGGCCCCTCCAAAAGTCTCATGTCCTCACATTATAAAACAATCATGCCTCCCAACAGTCCCGCATTAACCCAAAAGTCCGTAGTCCCGTATTAACTCAAAAGTCCACAGTCCAAAATCTCAATCTGAGACAAGGCAAGTTCCCTTCTGCCTATGAGACCTGTAAAATCAAAAGCAAGTTAATTACTTTCTAGATTACAAGGAGGTGAGATGTAAAATTAAAATGAAATAAGAAAAAGAGATGTGAAGTGGGGCTGATGTGAACTGAACATAGAGGGTTCATTTGTTAGAGGCCTGCAGTAACTTAAGGCTGATTTATGAGATATTTTTCTCCTTGTATTTTGATGAACATCAATAAGCCTCCTTGTATCTAGAAAGTAATTAACTCTTGCCTTTTGATTTTACAGGCTCATAGGCAGAAGGGACTTGCCTTGTCTCAGATGAGATTTTGGACTGTGGACTTTTGAGTTAATACGGGACTACGGACTTTTGGGTTAATGCGGGACTGTTGGAAGGCATGATTGGTTTTATAATGTGAGGACATGAGATTTGGAGGGCCAGGGGCCGTATGATATGGTTTGGCTGTCTCTCCACCCAAATATCAACTTGAATTGTATCTCCCAGAATTCTCATATGTTGTGGGAGGGACCCAGGGGGAGGTAATTGAATCATGGGGCCGTCTTTCCCATGCTATTCTCATGATAGTGAATAAGTCTCATGAGAGCTGATGGGTTTATCCAGGTTTCTGCTCTTGCTTCTTCCTCGTTTTCTCTTGCCGCCACCATGCAAGAAGGTGCCTTTCACCTCCCACCATGATTTGGATGCCTCCCCAGCCATGCAGAACTGTAAGTCCAATTAAACCACTTTTTCTTCCCAATCTTGGGGTGTGTCTTTATCAGCAGCATGAAAACAGACTAATACACAGGGCCGCATAAGGCACAATACAGAGAGATATTTTAAGGCAAGGCAGGAAGGCTAAGCTTTCATTTGATTCAGTGCTTTCAAATGTTCATCACCATCTAATATACCAGGGACAAATTATCAAAATTTTACATAATAATAAAAAGCATATGAAGATAAGAATGGTGGAAACAGATTCTGGAAAATCAACTAATAAATCATTAACATATCTCCAGATTCTGTCCAAAAAATATTGAAAAGATTCCTATCAGTCACAGTTCACATCAGCCCCACTTCACATCGCTTTTTCTTATTTCATTTTAATTTTACATCTCACCTCCTCATTCTTGCTTTAGATTCTGACAATTTCAGTGTGGTTAACAACTGTGACTGATAGGAATCTTTTCAATATTTTTTGGACAGAATCTGGAGATATGTTAATGATTTATTAGTTGATTTTCCAGAATCTGTTTCCACCATTCTTATCTTCATATGCTTTTTATTATTATGTAAAATTTTGATAATTTGTCCCTGGTATATTAGATGGTGATGAACATTTGAAAGCACTGAATCAAATGAAAGCTTAGCCTTCCTGCCTTGCCTTAAAATATCTCTCTGTATTGTGCCTTATGCGGCCCTGTGTATTAGTCTGTTTTCATGCTGCTGATAAAGACACACCCAAGATTGGGAAGAAAAAGTGGTTTAATTGGACTTACAGTTCTGCATGGCTGGGAGGCATCCAAATCATGGTGGGAGGTGAAAGGCACTTCTTGCATGGTGGCGGCAAGAGAAAACGAGGAAGAAGCAAGAGCAGAAACCTGGATAAACCCATCAGCTCTCATGAGACCTTATTCACTATCATGAGAATAAGCATGGGAAAGACCGGCCCCCATGATTCAATTACCTCCCCCTGGGTCCCTCCACAACATATGAGAATTCTGGGAGATACAATTCAAGTTGATATTTGGGTGGAGAGACAGCCAAACCATATCATACGGCCCCTGGCCCCTCCAAATCTCATGTCCTCACATTATAAAACCAATCATGCCTTCCAACAGTCCCGCATTAACCCAAAAGTCCGTAAGTCCCGTATTAACTCAAAAGTC
Seg_1 Overlap Distance chrX + 105093199 105093605 1 410 CGAGGAAGAAGCAAGAGCAGAAACCTGGATAAACCCATCAGCTCTCATGAGACTTATTCACTATCAATGAGAATAGCATGGAAAGACCGGCCCCCATGATTCAATTACCTCCCCCTGGGTCCCTCCCACAACATATGAGAATTCTGGAGATACAATTCAAGTTGATATTTGGGTGGAGAGACAGCCAAACCATATCATACGGCCCCTGGCCCCTCCAAAAGTCTCATGTCCTCACATTATAAAACAATCATGCCTCCCAACAGTCCCGCATTAACCCAAAAGTCCGTAGTCCCGTATTAACTCAAAAGTCCACAGTCCAAAATCTCAATCTGAGACAAGGCAAGTTCCCTTCTGCCTATGAGACCTGTAAAATCAAAAGCAAGTTAATTACTTTCTAGATTACAAGGAGG
Seg_2 Y:AGGAGG;AGGAGG:;:AGGAGG:6nt -1029 chrX - 105092746 105092582 405 570 AGGAGGTGAGATGTAAAATTAAAATGAAATAAGAAAAAGAGATGTGAAGTGGGGCTGATGTGAACTGAACATAGAGGGTTCATTTGTTAGAGGCCTGCAGTAACTTAAGGCTGATTTATGAGATATTTTTCTCCTTGTATTTTGATGAACATCAATAAGCCTCCTT
Seg_3 Y:CCTCCTT;CCTCCTT:;:CCTCCTT:7nt -1023 chrX - 105093605 105092787 564 1384 CCTCCTTGTATCTAGAAAGTAATTAACTCTTGCCTTTTGATTTTACAGGCTCATAGGCAGAAGGGACTTGCCTTGTCTCAGATGAGATTTTGGACTGTGGACTTTTGAGTTAATACGGGACTACGGACTTTTGGGTTAATGCGGGACTGTTGGAAGGCATGATTGGTTTTATAATGTGAGGACATGAGATTTGGAGGGCCAGGGGCCGTATGATATGGTTTGGCTGTCTCTCCACCCAAATATCAACTTGAATTGTATCTCCCAGAATTCTCATATGTTGTGGGAGGGACCCAGGGGGAGGTAATTGAATCATGGGGCCGTCTTTCCCATGCTATTCTCATGATAGTGAATAAGTCTCATGAGAGCTGATGGGTTTATCCAGGTTTCTGCTCTTGCTTCTTCCTCGTTTTCTCTTGCCGCCACCATGCAAGAAGGTGCCTTTCACCTCCCACCATGATTTGGATGCCTCCCCAGCCATGCAGAACTGTAAGTCCAATTAAACCACTTTTTCTTCCCAATCTTGGGGTGTGTCTTTATCAGCAGCATGAAAACAGACTAATACACAGGGCCGCATAAGGCACAATACAGAGAGATATTTTAAGGCAAGGCAGGAAGGCTAAGCTTTCATTTGATTCAGTGCTTTCAAATGTTCATCACCATCTAATATACCAGGGACAAATTATCAAAATTTTACATAATAATAAAAAGCATATGAAGATAAGAATGGTGGAAACAGATTCTGGAAAATCAACTAATAAATCATTAACATATCTCCAGATTCTGTCCAAAAAATATTGAAAAGATTCCTATCAGTCACAGTT
Seg_4 Y:CAGTT;CAGTT:;:CAGTT:5nt -930 chrX + 105092680 105093509 1380 2210 CAGTTCACATCAGCCCCACTTCACATCGCTTTTTCTTATTTCATTTTAATTTTACATCTCACCTCCTCATTCTTGCTTTAGATTCTGACAATTTCAGTGTGGTTAACAACTGTGACTGATAGGAATCTTTTCAATATTTTTTGGACAGAATCTGGAGATATGTTAATGATTTATTAGTTGATTTTCCAGAATCTGTTTCCACCATTCTTATCTTCATATGCTTTTTATTATTATGTAAAATTTTGATAATTTGTCCCTGGTATATTAGATGGTGATGAACATTTGAAAGCACTGAATCAAATGAAAGCTTAGCCTTCCTGCCTTGCCTTAAAATATCTCTCTGTATTGTGCCTTATGCGGCCCTGTGTATTAGTCTGTTTTCATGCTGCTGATAAAGACACACCCAAGATTGGGAAGAAAAAGTGGTTTAATTGGACTTACAGTTCTGCATGGCTGGGAGGCATCCAAATCATGGTGGGAGGTGAAAGGCACTTCTTGCATGGTGGCGGCAAGAGAAAACGAGGAAGAAGCAAGAGCAGAAACCTGGATAAACCCATCAGCTCTCATGAGACCTTATTCACTATCATGAGAATAAGCATGGGAAAGACCGGCCCCCATGATTCAATTACCTCCCCCTGGGTCCCTCCACAACATATGAGAATTCTGGGAGATACAATTCAAGTTGATATTTGGGTGGAGAGACAGCCAAACCATATCATACGGCCCCTGGCCCCTCCAAATCTCATGTCCTCACATTATAAAACCAATCATGCCTTCCAACAGTCCCGCATTAACCCAAAAGTCCGTAAGTCCCGTATTAACTCAAAAGTC
m64030_220717_044032/20/ccs 2888 TAGGGTGTCTGTACTCCAGCTCTGTCACCCAGGATAGAGTGCAATAGTGTGATCATAGCTCACTGCAACCTCAAACTTCTGGGCTCAAACAATCCTCCCACCTCAGTCCAGGGTTTTTCAGTCTGGCATGGAGATAAAGTAATGATCCCGTTCTCAATGACACATAGTTCAACTCTCCTGTTCCTCCTCAACTATTGATATCATCTTCCATCCCTCAATCCAAGAAATCTTTGTCTATTTCCAGGAATAAGAAAAATTGCCTAGCTGTATATTCTTCTAAATTAAGAAATTTTATTCCTGGGGTTTCAGCTTTTCAAACTCCAAAGTTAAGACTTGTACTCCTGCACTCTTAGTTGATACCAACTTTTCCAAATATTATGGGATAATACTGGGACGTATGAATTGGCAGCATACCACTGACCGAATGAGGATAAAGGCAAGAAATGCAAGGGAACAACAACAATGTAAAACACACCAGGTGGCCGGGTGCGGTGGCTCACACCTTGTAATCCCAGCATTTCCAGAGGCAAAGGCAGGAAGATCACTTGAGCACAGAAGTTCAAGATCAGCTCGGGCAATGTAGGGAGACCCCATTTCTACAAAACATAAAAAATATATTAGCCAAGTGTGGTAGCACATTACCTGTGGTCCCAACTACTCAGGATACTGAGGTGGGAAGATCACTTGAGCCCAGAAGGTTGAGGCTGCACTGAGCTGTGATCATGCCATTGCACTCTAGCCTGGGCAACAGAGTGAGACCCCATCTCAAAAAAAATAGTGTAATAAAACGCACCAGTTAATACATCAAAACATTATGCCACTTCTTTTTTTTGATCACTTAGGATGAAATGGATATGCCCCCTATAATAGGCTATGTCTGCACAATCATCTTATAATTGTTATCAGCCTGCCCATTGATCCAGAGTAAATTAGTCTGATGATATTTAAGTAAAACAGATTCTGTCCATAAAATTATGCAAAATGTAACAATGTTGACGCTGGTCATACTGTGCAAGCTAATAATTAGACATTGTCTATGTCATTTTCTGAACTTTCAGAGAGAACATTAAACAGTCAGACACTTGTTAATGGTAGAACATGGGGAAAATGACATGTGGTTTATGAATTTGGGGATTTTCCAGGGTTTTAGGAAGTCTCTGATAAGTTAAACAATCTATTCTTTAATGAGCTAACAGGGGGTTGGTATCTTCAAAAAACAATAGATGGAGATTAACCTATAAAAAAATTAATTAGAATATAAACACTGGCAGAATTTAAAGACAGAAAACTCTCTGTTCCCGTGGTTGAAGGATCCATTTCTTTGCTGTTTCCCGAAAAACTCCTTTCATGTTTCCTTCTCATCACTGTGTTCTGTTGATCCTTGCTCCTCAATTCTCCTGCCAGCCATTTTCCCTGGAAATGTTCTACCATTAACAAGTGTCTGACTGTTTAATGTTCTCTCTGAAAGTTCAGAAAATGACATAGACAATGTCTAATTATTAGCTTGCACAGTATGACCAGCGTCAACATTGTTACATTTTGCATAATTTTATGGACAGAATCTGTTTTACTTAAATATCATCAGACTAATTTACTCTGGATCAATGGGCAGGCTGATAACAATTATAAGATGATTGTGCAGACATAGCCTATTATAGGGGGCATATCCATTTCATCCTAAGTGATCAAAAAAAGAAGTGGCATAATGTTTTGATGTATTAACTGGTGCGTTTTATTACACTATTTTTTGAGATGGGGTCTCACTCTGTTGCCCAGGCTAGAGTGCAATGGCATGATCACAGCTCAGTGCAGCCTCAACCTTCTGGGCTCAAGTGATCTTCCCACCTCAGTATCCTGAGTAGTTGGGACCACAGGTAATGTGCTACCACACTTGGCTAATATATTTTTTATGTTTTGTAGAAATGGGGTCTCCCTACATTGCCCGAGCTGATCTTGAACTTCTGTGCTCAAGTGATCTTCCTGCCTTTGCCTCTGGAAATGCTGGGATTACAAGGTGTGAGCCACCGTACCCGGCCACCTGGTGTGTTTTACATTGTTGTTGTTCCCTTGCATTTCTTGCCTTTATCCTCATTCGGTCAGTGGTATGCTGCCAATTCATACGTCCCAGTATTATCCCATAATATTTGGAAAAGTTGGTATCAACTAAGAGTGCAGGAGTACAAGTCTTAACTTTGGAGTTTGAAAAGCTGAAACCCCAGGAATAAAATTTCTTAATTTAGAAGAATATACAGCTAGGCAATTTTTCTTATTCCTGGAAATAGACAAAGATTTCTTGGATTGAGGGATGGAAGATGATATCAATAGTTGAGGAGGAACAGGAGAGTTGAACTATGTGTCATTGAGAACGGGATCATTACTTTATCTCCATGCCAGACTGAAAAACCCTGGACTGGGGATGAAATCAGATAAAAAAAAATTCTGGTAATTTTGGGAAATGTGAAAACAGAGAAATTTTGCCCTGTGCTCCCTGGATAGAACCCAAGAAAGCAGCAAAATCTCAGGTTGCTTGCCCTGTGGGGATTGTATTTAATCATTCACACACTGATTTGTCTGTTCAACAGGTATTTTCTTTTTCATTGATTGATGGATTGATTGAGAAAGGGTGTCTGTACTCCAGCTCTGTCACCCAGGATAGAGTGCAATAGTGTGATCATAGCTCACTGCAACCTCAAACTTCTGGGCTCAAACAATCCTCCCACCTCAGTCCAGGGTTTTTCAGTCTGGCATGGAGATAAAGTAATGATCCCGTTCTCAATGACACATAGTTCAACTCTCCTGTTCCTCCTCAACTATTGATATCATCTTCCATCCCTCAATCCAAGAAATCTTTGTCTATTTCCAGGAATAAGAAATTGTGCTGTAATTTA
Seg_1 Overlap Distance chr10 - 16374976 16374870 2 108 AGGGTGTCTGTACTCCAGCTCTGTCACCCAGGATAGAGTGCAATAGTGTGATCATAGCTCACTGCAACCTCAAACTTCTGGGCTCAAACAATCCTCCCACCTCAGTC
Seg_2 Y:CAGTC;CAGTC:;:CAGTC:5nt 203 chr10 + 16375184 16376188 104 1110 CAGTCCAGGGTTTTTCAGTCTGGCATGGAGATAAAGTAATGATCCCGTTCTCAATGACACATAGTTCAACTCTCCTGTTCCTCCTCAACTATTGATATCATCTTCCATCCCTCAATCCAAGAAATCTTTGTCTATTTCCAGGAATAAGAAAAATTGCCTAGCTGTATATTCTTCTAAATTAAGAAATTTTATTCCTGGGGTTTCAGCTTTTCAAACTCCAAAGTTAAGACTTGTACTCCTGCACTCTTAGTTGATACCAACTTTTCCAAATATTATGGGATAATACTGGGACGTATGAATTGGCAGCATACCACTGACCGAATGAGGATAAAGGCAAGAAATGCAAGGGAACAACAACAATGTAAAACACACCAGGTGGCCGGGTGCGGTGGCTCACACCTTGTAATCCCAGCATTTCCAGAGGCAAAGGCAGGAAGATCACTTGAGCACAGAAGTTCAAGATCAGCTCGGGCAATGTAGGGAGACCCCATTTCTACAAAACATAAAAAATATATTAGCCAAGTGTGGTAGCACATTACCTGTGGTCCCAACTACTCAGGATACTGAGGTGGGAAGATCACTTGAGCCCAGAAGGTTGAGGCTGCACTGAGCTGTGATCATGCCATTGCACTCTAGCCTGGGCAACAGAGTGAGACCCCATCTCAAAAAAAATAGTGTAATAAAACGCACCAGTTAATACATCAAAACATTATGCCACTTCTTTTTTTTGATCACTTAGGATGAAATGGATATGCCCCCTATAATAGGCTATGTCTGCACAATCATCTTATAATTGTTATCAGCCTGCCCATTGATCCAGAGTAAATTAGTCTGATGATATTTAAGTAAAACAGATTCTGTCCATAAAATTATGCAAAATGTAACAATGTTGACGCTGGTCATACTGTGCAAGCTAATAATTAGACATTGTCTATGTCATTTTCTGAACTTTCAGAGAGAACATTAAACAGTCAGACACTTGTTAATGGTAGAACATGGGGAAAATG
Seg_3 Y:CATGGGGAAAATG;CATGGGGAAAATG:;:CATGGGGAAAATG:13nt -1331 chr10 - 16376494 16374870 1098 2727 CATGGGGAAAATGACATGTGGTTTATGAATTTGGGGATTTTCCAGGGTTTTAGGAAGTCTCTGATAAGTTAAACAATCTATTCTTTAATGAGCTAACAGGGGGTTGGTATCTTCAAAAAACAATAGATGGAGATTAACCTATAAAAAAATTAATTAGAATATAAACACTGGCAGAATTTAAAGACAGAAAACTCTCTGTTCCCGTGGTTGAAGGATCCATTTCTTTGCTGTTTCCCGAAAAACTCCTTTCATGTTTCCTTCTCATCACTGTGTTCTGTTGATCCTTGCTCCTCAATTCTCCTGCCAGCCATTTTCCCTGGAAATGTTCTACCATTAACAAGTGTCTGACTGTTTAATGTTCTCTCTGAAAGTTCAGAAAATGACATAGACAATGTCTAATTATTAGCTTGCACAGTATGACCAGCGTCAACATTGTTACATTTTGCATAATTTTATGGACAGAATCTGTTTTACTTAAATATCATCAGACTAATTTACTCTGGATCAATGGGCAGGCTGATAACAATTATAAGATGATTGTGCAGACATAGCCTATTATAGGGGGCATATCCATTTCATCCTAAGTGATCAAAAAAAGAAGTGGCATAATGTTTTGATGTATTAACTGGTGCGTTTTATTACACTATTTTTTGAGATGGGGTCTCACTCTGTTGCCCAGGCTAGAGTGCAATGGCATGATCACAGCTCAGTGCAGCCTCAACCTTCTGGGCTCAAGTGATCTTCCCACCTCAGTATCCTGAGTAGTTGGGACCACAGGTAATGTGCTACCACACTTGGCTAATATATTTTTTATGTTTTGTAGAAATGGGGTCTCCCTACATTGCCCGAGCTGATCTTGAACTTCTGTGCTCAAGTGATCTTCCTGCCTTTGCCTCTGGAAATGCTGGGATTACAAGGTGTGAGCCACCGTACCCGGCCACCTGGTGTGTTTTACATTGTTGTTGTTCCCTTGCATTTCTTGCCTTTATCCTCATTCGGTCAGTGGTATGCTGCCAATTCATACGTCCCAGTATTATCCCATAATATTTGGAAAAGTTGGTATCAACTAAGAGTGCAGGAGTACAAGTCTTAACTTTGGAGTTTGAAAAGCTGAAACCCCAGGAATAAAATTTCTTAATTTAGAAGAATATACAGCTAGGCAATTTTTCTTATTCCTGGAAATAGACAAAGATTTCTTGGATTGAGGGATGGAAGATGATATCAATAGTTGAGGAGGAACAGGAGAGTTGAACTATGTGTCATTGAGAACGGGATCATTACTTTATCTCCATGCCAGACTGAAAAACCCTGGACTGGGGATGAAATCAGATAAAAAAAAATTCTGGTAATTTTGGGAAATGTGAAAACAGAGAAATTTTGCCCTGTGCTCCCTGGATAGAACCCAAGAAAGCAGCAAAATCTCAGGTTGCTTGCCCTGTGGGGATTGTATTTAATCATTCACACACTGATTTGTCTGTTCAACAGGTATTTTCTTTTTCATTGATTGATGGATTGATTGAGAAAGGGTGTCTGTACTCCAGCTCTGTCACCCAGGATAGAGTGCAATAGTGTGATCATAGCTCACTGCAACCTCAAACTTCTGGGCTCAAACAATCCTCCCACCTCAGTC
Seg_4 Y:CAGTC;CAGTC:;:CAGTC:5nt -1315 chr10 + 16375184 16375334 2723 2873 CAGTCCAGGGTTTTTCAGTCTGGCATGGAGATAAAGTAATGATCCCGTTCTCAATGACACATAGTTCAACTCTCCTGTTCCTCCTCAACTATTGATATCATCTTCCATCCCTCAATCCAAGAAATCTTTGTCTATTTCCAGGAATAAGAAAThe overlap information represented the overlap sequence (reverse extension of overlapping segment) and overlap length between two adjacent segments. The last two strings is the overlap sequence and overlap length.
This script ChimerasDownstream.pl is used for downstream analysis of chimeras:
-
Merge chimeras of each sub-chimera file to a file
-
Tranforming the raw format of chimeras to a better format for viewing
-
Extracting the direct chimera and inverted chimera to different files.
-
Count the chimera types'++' '--' '+-' '-+' , and the number of direct chimera , inverted chimera, chimera's number, chimeric site and total length of chimeras.
The usage of it is following as:
perl ChimerasDownstream.pl
Usage: perl ChimerasDownstream.pl
For Extracting, transformating, and getinfo of chimeras.
-d <string> <the directory of all chimeras file>
-od <string> <the directory for saving the results> <dflt = $dir/chimeras_analysis_lap(the min overlap length)>
-n <string> <the name of this sample>
-L <INT> <min length of segment> <dflt = 50>
-p <INT> <the min overlap length> <dflt = 3>
-s <INT> <the smallest distance of two segment> <dflt = 25>
-b <INT> <the biggest distance of two segment> <dflt = 10000>
Example: perl ChimerasDownstream.pl -d /home/luna/work/ThirdChimera/0907data/5e.2 -n 5e.2 -L 50 -s 25 -b 10000
The example:
# Here, we assume that the length threshold of the overlapping sequence was set to 2 nt.
perl $dir/ChimerasDownstream.pl -d $dir/exampledata/result/$samp -od $dir/exampledata/result/$samp/chimeras_analysis_lap2 -n $samp -p 2 This script SplitChimericRead.pl is used to split the chimeric read into multiple normal sgements. The usage of it is following as:
perl SplitChimericRead.pl
Usage:
perl SplitChimericRead.pl [options]
Options:
-i <file> <in.input raw bam file>
-f <file> <input id file>
-c <file> <input chimera file>
-o <string> <out.output fastq file>The example:
# Here, we assume that the length threshold of the overlapping sequence was set to 2 nt.
perl $dir/SplitChimericRead.pl -i $dir/exampledata/$samp.ccs.bam -f $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.chimera.id -c $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.normal.txt -o $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.chimera.fqUse the script LapLengthGC.pl for counting:
# for all chimeras
perl $dir/LapLengthGC.pl -i $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.normal.txt -o $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.LapGC.txt -n $samp
# for inverted chimeras
perl $dir/LapLengthGC.pl -i $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.inverted -o $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.IV.LapGC.txt -n $samp
# for direct chimeras
perl $dir/LapLengthGC.pl -i $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.direct -o $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.DR.LapGC.txt -n $sampUse the script DistanceCount.pl for counting:
# for all chimeras
perl $dir/DistanceCount.pl -i $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.normal.txt -d $dir/exampledata/result/$samp/chimeras_analysis_lap2 -o $samp.ALL -sl 5
# for inverted chimeras
perl $dir/DistanceCount.pl -i $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.inverted -d $dir/exampledata/result/$samp/chimeras_analysis_lap2 -o $samp.IV -sl 5
# for direct chimeras
perl $dir/DistanceCount.pl -i $dir/exampledata/result/$samp/chimeras_analysis_lap2/$samp.direct -d $dir/exampledata/result/$samp/chimeras_analysis_lap2 -o $samp.DR -sl 5see the details in the GitHub Page.
We will be pleased to address any question or concern you may have with the 3rd-ChimeraMiner: [email protected]
Lu, N., Qiao, Y., An, P., Luo, J., Bi, C., Li, M., Lu, Z., & Tu, J. Exploration of whole genome amplification generated chimeric sequences in long-read sequencing data, Briefings in Bioinformatics, 2023, bbad275, https://doi.org/10.1093/bib/bbad275.